Fig. 5.
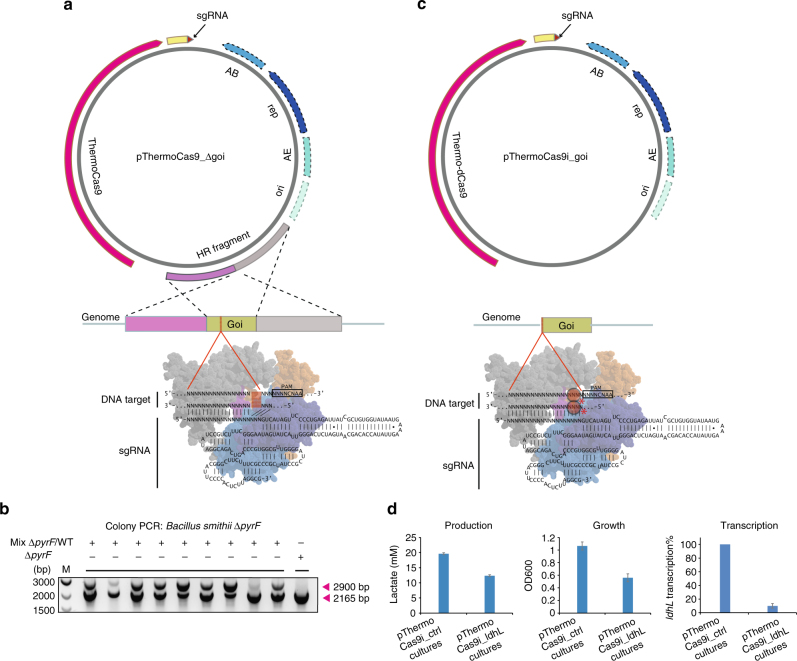
ThermoCas9-based genome engineering in prokaryotes. a Schematic overview of the basic pThermoCas9_Δgene-of-interest (goi) construct. The thermocas9 gene was introduced either to the pNW33n (B. smithii) or to the pEMG (P. putida) vector. Homologous recombination flanks were introduced upstream thermocas9 and encompassed the 1 kb (B. smithii) or 0.5 kb (P. putida) upstream and 1 kb or 0.5 kb downstream region of the gene of interest (goi) in the targeted genome. A sgRNA-expressing module was introduced downstream the thermocas9 gene. As the origin of replication (ori), replication protein (rep), antibiotic resistance marker (AB) and possible accesory elements (AE) are backbone specific, they are represented with dotted outline. b Agarose gel electrophoresis showing the resulting products from genome-specific PCR on ten colonies from the ThermoCas9-based pyrF deletion process from the genome of B. smithii ET 138. All ten colonies contained the ΔpyrF genotype and one colony was a clean ΔpyrF mutant, lacking the wild-type product. Supplementary Fig. 13 shows the uncropped gel image. c Schematic overview of the basic pThermoCas9i_goi construct. Aiming for the expression of a catalytically inactive ThermoCas9 (ThermodCas9: D8A, H582A mutant), the corresponding mutations were introduced to create the thermodcas9 gene. The thermodcas9 gene was introduced to the pNW33n vector. A sgRNA-expressing module was introduced downstream the thermodcas9. d Graphical representation of the production, growth and RT-qPCR results from the ldhL silencing experiment using ThermodCas9. The graphs represent the lactate production, optical density at 600 nm and percentage of ldhL transcription in the repressed cultures compared to the control cultures. Average values from three biological replicates are shown, with error bars representing S.D