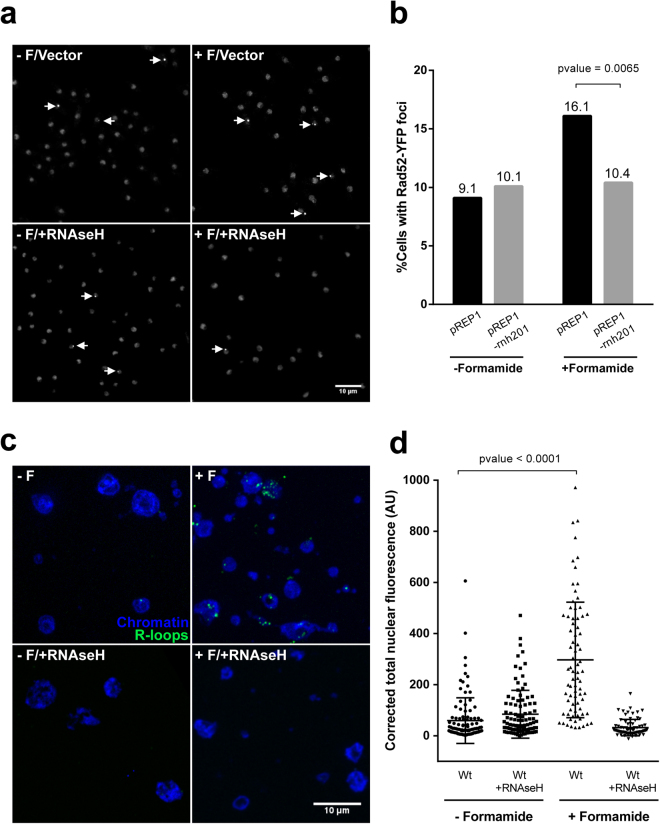
Figure 6.
Formamide enhances R-loop formation. (a) Examples of average intensity projections of 10 slices stacks from rad52-YFP cells in all four combinations: bearing either pRep1 empty expression vector (upper panels) or over-expressing RNAse H catalytic subunit (Rnh201) under nmt1 promoter (lower panels) and in the absence (left panels) or the presence (right panels) of formamide for three hours. Arrows indicate nuclei containing Rad52 foci. Scale bar: 10 µm. (b) Percentage of cells containing Rad52 foci in each condition (n ≥ 520). p-value was determined using a two-sided Chi-square test. (c) Chromosome spreads immunofluorescence. Examples of merged DAPI (chromatin)/Alexa488 (R-loops) projections (10 slices stacks) from wild type cells in all four combinations: no formamide no RNAse H (upper left panel), plus formamide no RNAse H (upper right panel), no formamide plus RNAse H (lower left panel), plus formamide plus RNAse H (lower right panel). Scale bar: 10 µm. (d) Fluorescent signal quantification in nuclear spreads. Integrated fluorescent density from each nucleus was made relative to the size and field’s average background signal subtracted. Resulting intensity (arbitrary units) is represented for at least 75 nuclei on each condition. p-value was determined using a t-test.