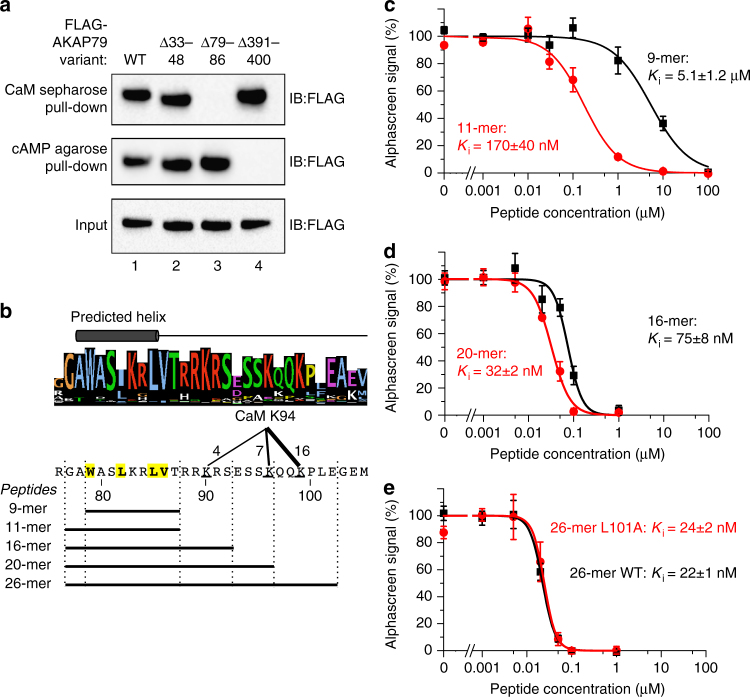
Fig. 3.
Delineation of key residues in AKAP79 required for CaM binding. a Pull-down of either WT, Δ33–48, Δ79–86, or Δ391–400 FLAG-tagged-AKAP79 (inputs shown in bottom panel) with either CaM sepharose (top panel) or cAMP agarose (middle panel). AKAP79 was detected by anti-FLAG immunoblotting. The experiment was performed in triplicate with each replicate producing the same pattern of bands. b Sequence LOGO for AKAP5 gene products aligned with predicted helical region. The cross-linking cluster between AKAP79 positions 90–99 and K94 in CaM is indicated along with the boundaries of peptides used in the following panels. c–e Determination of inhibitory constants for the peptides outlined in (b) in disrupting interaction between biotin-CaM and GST-AKAP79 (1–153) detected using the alphascreen assay (n = 4 for all data points). Ki constants were determined for the 9-mer and 11-mer peptides c, 16-mer and 20-mer peptides d, and for either WT or L101A 26-mer peptides e.***P<0.001 by two-tailed Student's t-test. Error bars show s.e.m.