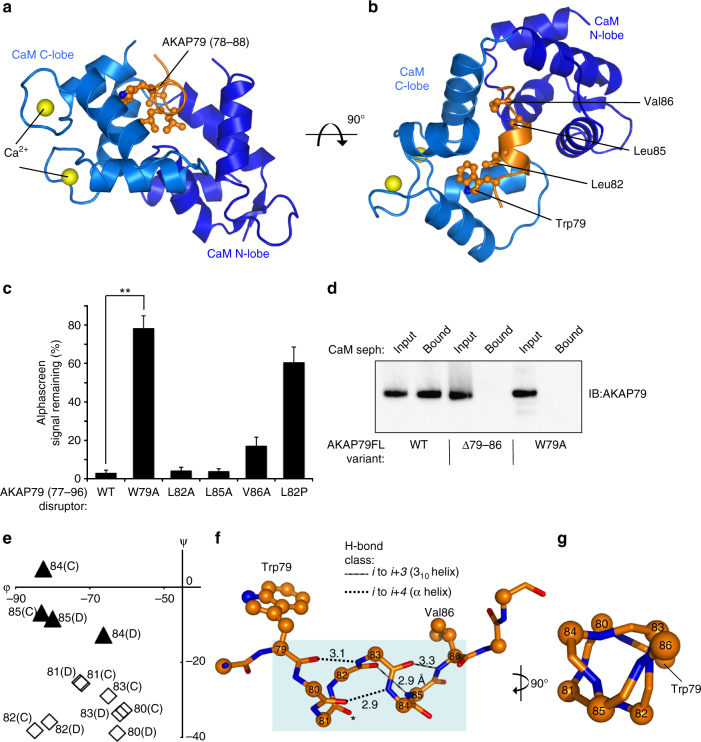
Fig. 5.
Crystal structure of CaM in complex with its AKAP79 binding site. a Cartoon representation showing one of the two copies (chains B and D) of AKAP79 peptide (orange) bound to CaM (blue) in the asymmetric unit. The C-lobe (lighter blue) is in the open conformation with each of its two EF hands coordinating Ca2+ (yellow). b Rotation of the complex through 90° highlighting the position of the four hydrophobic amino acids comprising the 1-4-7-8 motif. c Reduction in alphascreen signal between biotin-CaM and GST-AKAP79 (1–153) upon addition of 20-mer peptides derived from AKAP79 77–96 (n = 4). The effects of point mutations within the disruptor peptide were compared. d Binding of purified full-length WT, Δ79–86 or W79A AKAP79 to CaM sepharose. Each AKAP79 variant was purified in complex with the D/D of RIIα. AKAP79 was released from the beads by incubation with EGTA, and detected by anti-AKAP79 IB. The experiment was performed in triplicate with each replicate leading to the same pattern of bands. e Limited Ramachandran plot showing dihedral angles for both copies of AKAP79 positions 80–85 in the asymmetric unit. Black triangles represent amino acids with angles characteristic of 310 helices; white diamonds are amino acids with α-helical geometry. f Representation of backbone H-bonds within the AKAP79 helix with distances shown in Å. The two α-helix-type bonds are shown by dotted lines; 310-helical H-bonds as striped lines. The carbonyl group of S81 that does not H-bond to a backbone group is asterisked. g Rotation of the helix through 90°. The triangular backbone geometry of positions 83–86 is such that the side-chains of W79, L83 and T86 extend in the same direction. **P < 0.01 by two-tailed Student’s t test. Error bars show s.e.m.