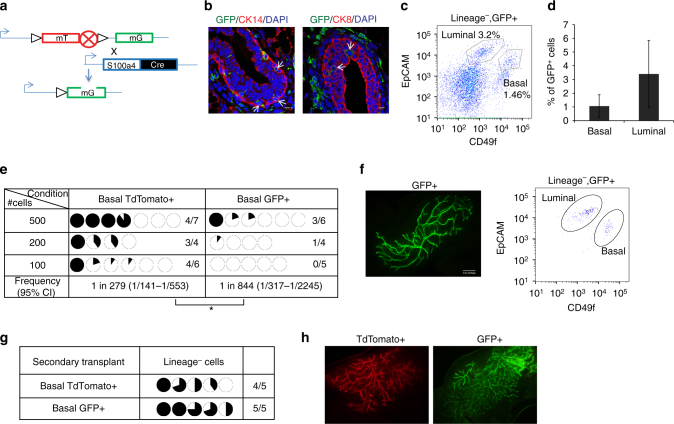
Fig. 2.
S100a4 lineage traced basal cells can regenerate the mammary gland in vivo. a Schematic diagram showing the crossing of Rosa26 mTmG reporter with S100a4-cre mice. b Immunofluorescence analysis showing that S100a4 lineage traced cells (GFP+) are present in mammary epithelial cells. CK8/CK14 (red), GFP (green), and DAPI (blue). Arrows indicate S100a4 traced epithelial cells. Scale bar, 10 µm. c Representative FACS analysis of the S100a4 cre /Rosa26 mTmG mouse mammary gland of Lineage−/Live−/GFP+. (n = 20 mice). Also see Supplementary Fig. 2a. d Percentage of GFP+ cells in basal and luminal cell populations. The data are shown as mean ± SD (n = 10 mice). e Basal TdTomato+ and GFP+ cells were sorted from 10–12-week-nulliparous females and transplanted in 3-week old mice. All transplants were analyzed after 8 weeks. The data are combined from three independent transplantation experiments. Pie charts show the percentage of mammary fat pad filled. (*p < 0.05, frequencies and p-value are calculated using ELDA software). Also see Supplementary Fig. 2 a for sorting scheme. f Representative image of a basal GFP+ transplant (left). FACS analysis using EpCAM and CD49f showing basal and luminal cell populations (right) (n = 3 outgrowths). g Secondary transplant of cells from e. Pie charts show percentage of mammary fat pad filled. h Representative image of secondary transplants from TdTomato+ (left) and GFP+ basal cells (right) (n = 4 outgrowths)