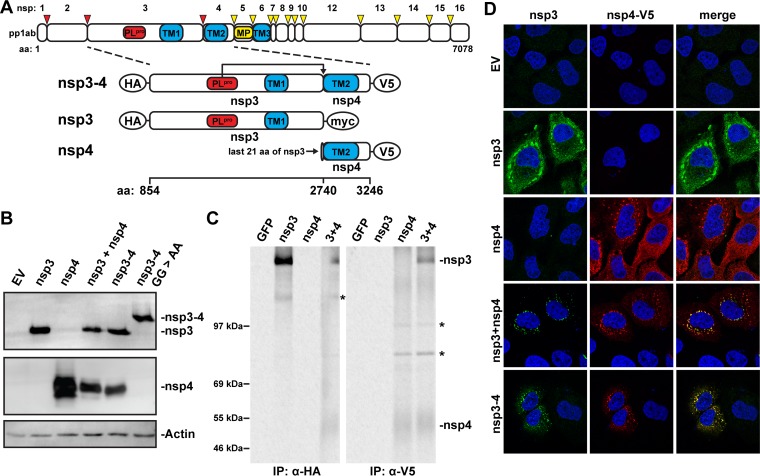
FIG 1 .
MERS-CoV nsp3 and nsp4 interact with each other. (A) Scaled schematic overview of MERS-CoV pp1ab and nsp3-4 constructs. Amino acid numbers refer to the MERS-CoV pp1ab sequence. The expected cleavage of the nsp3/nsp4 junction by PLpro is indicated. The epitope tags used at the termini of the constructs are indicated with ovals. TM, transmembrane region. (B) 293T cells were transfected with MERS-CoV nsp3-4 plasmids or empty pCAGGS vector (EV) and analyzed by Western blotting 20 h posttransfection. nsp3 was detected with anti-SARS-CoV nsp3 serum that cross-reacts with MERS-CoV nsp3 (21), and nsp4 was detected with anti-V5 monoclonal antibody. (C) Constructs expressing MERS-CoV nsp3 or nsp4 or a GFP control were transfected into 293T cells, which were metabolically labeled with [35S]methionine-cysteine from 4 to 20 h posttransfection. Lysates were immunoprecipitated with the indicated antibodies, separated on an SDS-PAGE gel, and visualized using phosphorimaging. Bands not corresponding to expected protein size in the Western blot are indicated with asterisks. The ~130-kDa band in the nsp3 IP was also observed in the Western blot. nsp4 bands in IP were fuzzy likely due to the relatively high hydrophobicity of the protein. (D) HuH-7 cells were transfected with the indicated plasmids, and localization of MERS-CoV nsp3 and nsp4 was analyzed using immunofluorescence labeling and confocal microscopy at 24 h posttransfection. nsp3 was detected with anti-SARS-CoV-nsp3 serum, and nsp4 was detected with anti-V5 monoclonal antibody.