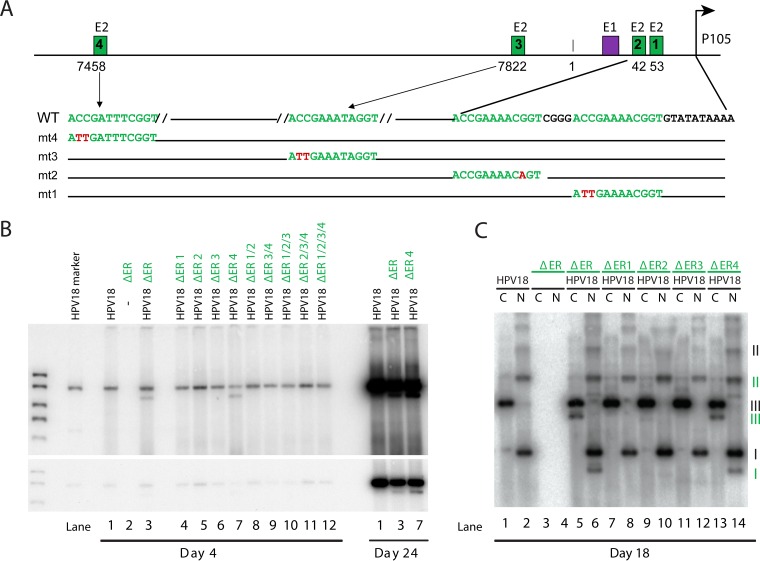
FIG 2 .
E2BS4 is not required for long-term genome maintenance. (A) Diagram of the positions and sequences of E2 binding sites in the HPV18 URR. Shown is the position of the core origin of replication, the constitutive enhancer, E1 and E2 binding sites, and the TATA box and position of the P105 early promoter. Nucleotide substitutions generated in each binding site are shown in red and labeled mt1 to -4, corresponding to E2BS1 to -4. Previous studies have shown that these mutations abrogate E2 binding (26). (B) ΔER replicons containing the E2BS mutated alone or in combination (e.g., ΔER 1/2 has mutations in E2BS1 and -2 [mt1 and mt2, as shown in panel A]) were cotransfected with the HPV18 genome into primary HFKs. pUC18 was used to balance the amount of transfected DNA. Low-molecular-weight DNA was extracted at the times shown, digested with DpnI and a linearizing enzyme, and then analyzed by Southern blotting. Two exposures of a representative blot are shown (n = 2, with different collection times). (C) Low-molecular-weight DNA from a subset of the transfected cells shown in panel B was harvested from cells at 18 days posttransfection and digested with an enzyme that either linearized (C) or did not cut (N) the HPV18 genome (and derived replicons). DNA was analyzed using Southern blotting. The different forms of extrachromosomal DNA are indicated (black roman numerals indicate the wild-type genome; subgenomic replicons are represented by green numerals). I, supercoiled; II, nicked circle; III, linear. n = 2, with different collection times.