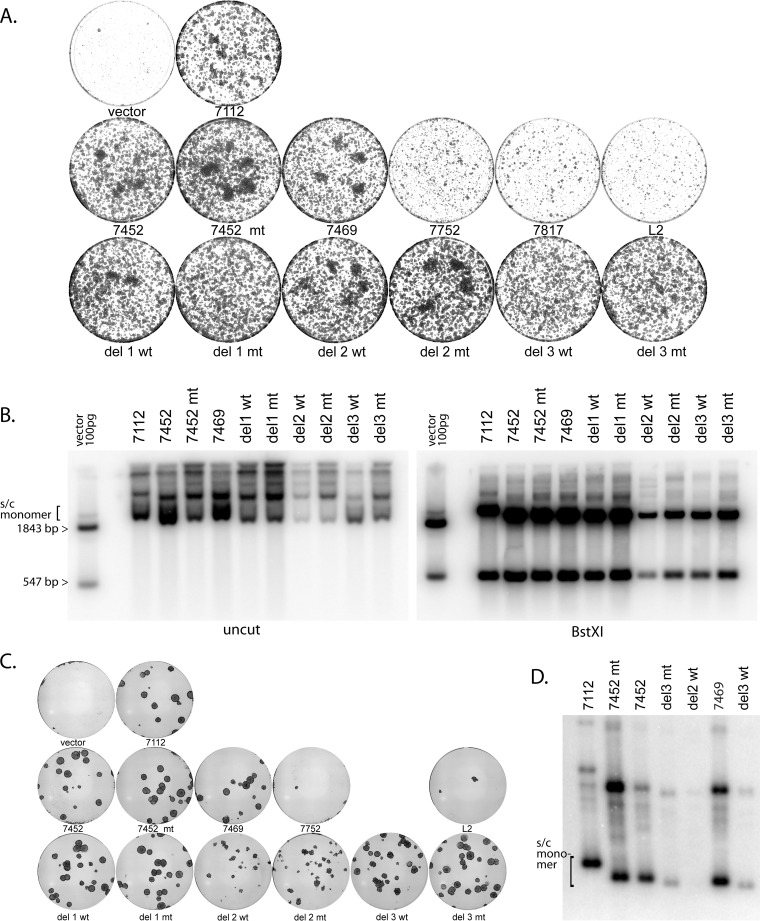
FIG 5 .
Quantitative colony formation assay results, showing that the viral replication origin and the viral enhancer are required for efficient long-term maintenance. (A) Cells were cotransfected with the HPV18 genome and the series of replicons shown in Fig. 3 and then selected with G418 at 400 µg/ml for 2 days and then 200 µg/ml until visible colonies formed (8 to 9 days posttransfection). Colonies were stained with methylene blue and imaged. The plasmid URR fragment nomenclature is as shown in Fig. 3. (B) Cell DNA was prepared from pooled colonies from a duplicate set of plates and was analyzed using Southern blotting. The samples on the left gel were cleaved with NdeI, which does not cut the pCpG neo URR plasmids; for the right gel, they were cleaved with BstXI. See Fig. 3A for the positions of restriction enzyme sites. The exact experiment shown is representative of two independent experiments. Multiple other replicates (with subsets of plasmids and/or different timings) gave comparable results. (C) Cells were transfected and selected as described for panel A until large colonies were apparent (15 days posttransfection). Colonies were stained and imaged (A) or pooled prior to DNA extraction (D). (D) Cellular DNA was prepared from those colonies that survived selection, and equal amounts were analyzed by Southern blotting. DNA was cut with a restriction enzyme that did not cleave the replicons. The position of supercoiled monomers is indicated.