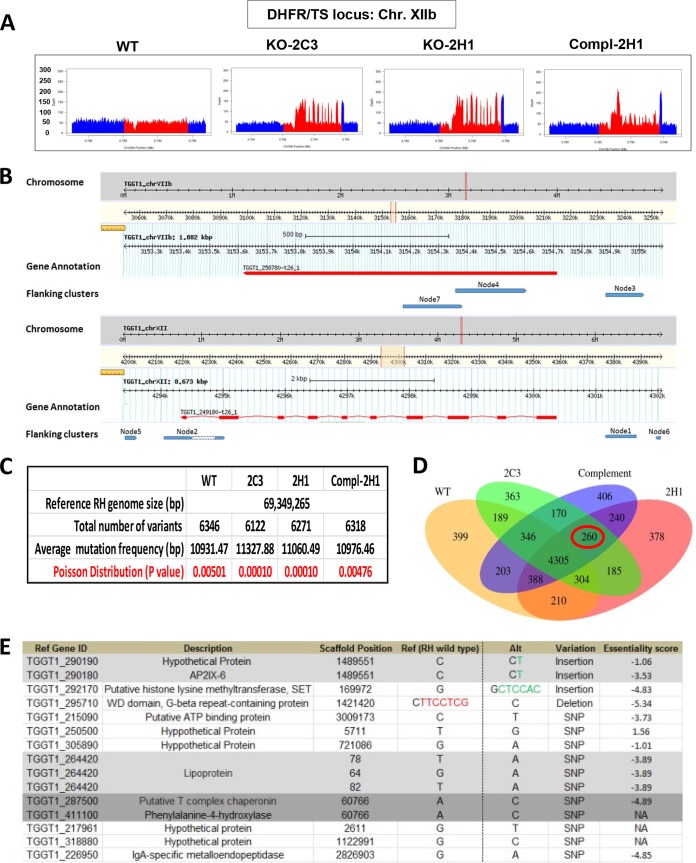
FIG 9 .
Whole-genome next-generation sequencing of 4 parasite lines (WT, 2 KOs, and a complemented line). (A) Each chart shows the read depth coverage of a portion of the T. gondii chromosome XIIb, where the red region represents the DHFR/TS locus, showing 3 copies of DHFR in the TgOTUD3A-KO samples (clones 2C3 and 2H1) and 2 copies of DHFR in the complemented 2H1 (Compl-2H1 [clone D5]) samples, compared to 1 in the wild type. (B) Analysis of the locations of the extra DHFR copies in the TgOTUD3A-KO and complemented lines. Flanking sequences of the drug cassettes in the top panel of the genome browser (derived from ToxoDB) output represent the TgOTUD3A locus, where 3 of the 7 hits match, and the bottom panel shows the DHFR locus, where the remaining 4 of the 7 hits aligned, indicating that the drug cassettes integrated in tandem in the TgOTUD3A locus without random insertion in the genome. (C) Statistical analysis shows that the average frequencies of mutation (based on the total number of variations and RH genome size) found in each sample and between samples are random and not caused by CRISPR editing. (D) Venn diagram comparing all variants called by our analysis, showing that 260 of them (circled in red) were shared between the mutants and not with the wild type. (E) Further analysis determined that out of 260 variants, only 15 were found inside the coding regions; these are listed in the table. Since there is no RH annotation available in the database, we used the GT1 gene identification number (ID) as the reference gene. Position, type of mutation, and essentiality score based on a recent genome-wide CRISPR study (38) are provided.