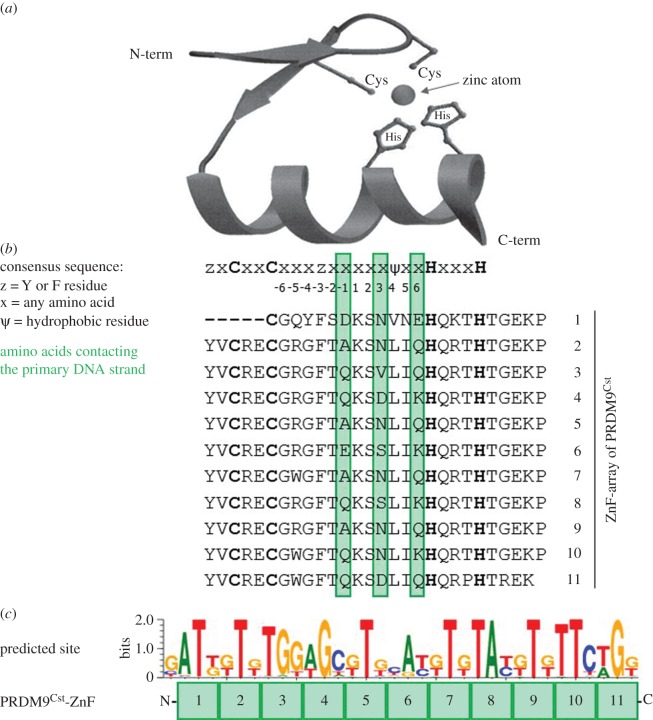
Figure 1.
(a) Three-dimensional structure of a representative single C2H2-type ZnF, harbouring two antiparallel beta-sheets and an alpha helix, which contains the DNA-binding amino acids, as well as a zinc atom coordinated by two cysteines and two histidines (adapted from Wolfe et al. [68]). (b) Consensus sequence of a C2H2-type ZnF [68] aligned with the ZnF array of PRDM9Cst. The DNA-contacting residues (at positions −1, 3 and 6) are highlighted in green. The zinc coordinating amino acids are shown in bold letters. (c) Predicted binding site of the 11 PRDM9Cst-ZnFs using the Persikov algorithm (polynomial SVM settings) [69] aligned to the respective ZnFs. Shown is the reverse DNA strand in 5′–3′ direction (the amino acids at positions −1, 3 and 6 contact the forward strand).