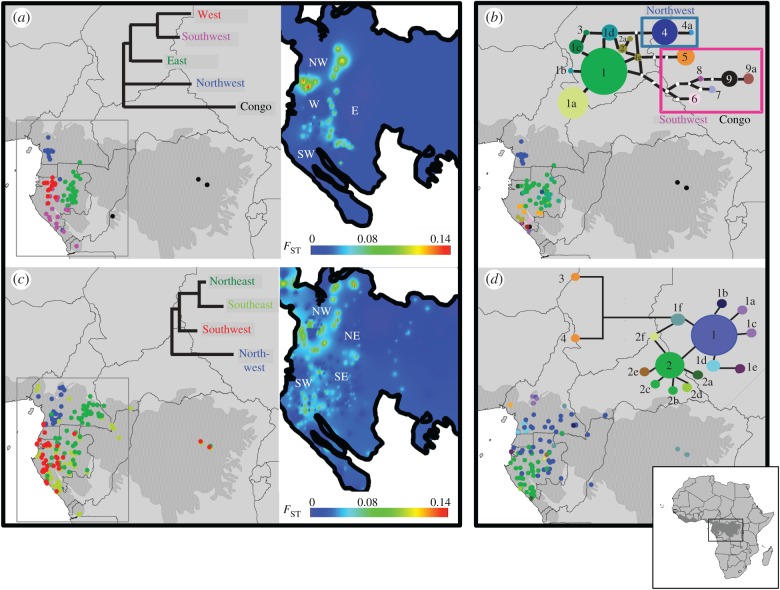
Figure 3.
(a,c) Geographical distribution and genetic distances among nSSR Bayesian genetic clusters of (a) S. zenkeri (10 loci, 240 individuals) and (c) G. suaveolens (8 loci, 450 individuals). Only non-admixed individuals, i.e. showing more than 70% ancestry in the Bayesian clustering, are represented. Neighbour-joining trees were built from pairwise FST among nSSR genetic clusters. Heat maps represent interpolation surfaces of FST implemented with SPADS. (b,d) Geographical distribution and genealogy of trnC-petN1R for (b) S. zenkeri (17 haplotypes, 192 individuals) and (d) G. suaveolens (16 haplotypes, 260 individuals).