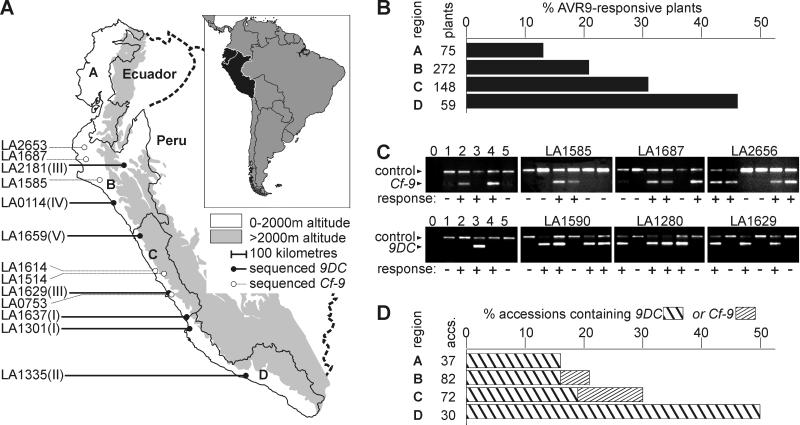
Figure 5.
Frequency of AVR9 recognition and occurrence of 9DC and Cf-9 in the Lp population. (A) The natural distribution range of Lp. The Lp distribution range is bordered by the Pacific Ocean in the west and the 2,000-m elevation line of the Andes Mountains in the east. This area is divided into four regions: Ecuador (A) and northern (B), central (C), and southern (D) Peru. Accessions from which entire 9DC or Cf-9 ORFs have been sequenced are indicated on the left, with allelic classes I–V between brackets (see Fig. 6). (B) Frequency of AVR9-responsive plants per region. For each region, the number of AVR9-responsive plants was divided by the total number of AVR9-injected plants originating from that region. Not all plants could be mapped to regions. (C) Amplification of fragments of Cf-9 and 9DC. Specific primers were tested (lanes 0–5) and used to detect the presence of 9DC and Cf-9 genes in Lp accessions that contain both AVR9-responsive and nonresponsive plants (panels marked with LA numbers). Specific amplification products of Cf-9, 9DC, and Act (control) genes were obtained as explained in Materials and Methods. Templates were genomic DNA isolated from the following sources: 1, MoneyMaker (MM)-Cf0 tomato; 2, MM-Cf9 tomato; 3, AVR9-responsive Lp plant of accession LA1301; 4, Cf-9-transgenic MM-Cf0 tomato; and 5, Hcr9-9D-transgenic MM-Cf0 tomato. Water (lane 0) was used as a negative control. AVR9 responsiveness is indicated by − or + below the panels. (D) Frequency of 9DC and Cf-9 genes per region. One AVR9-responsive plant of each accession was analyzed for the presence of 9DC or Cf-9 genes. The number of accessions that contained 9DC or Cf-9 was divided by the total number of accessions from that region. The frequency has been adjusted for the number of plants tested per accession. Not all accessions could be mapped to regions.