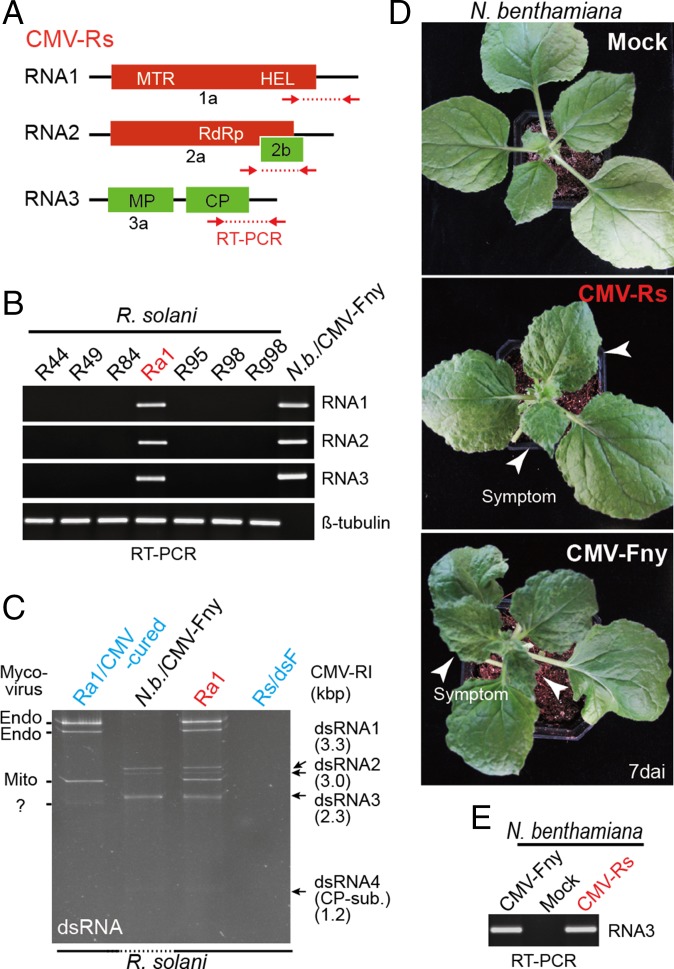
Fig. 1.
CMV infection in R. solani. (A) Schematic representation of genome structure of CMV-infecting R. solani (not to scale). Colored boxes and lines indicate the reading frame (ORF) and untranslated regions (UTR), respectively. (B and E) RT-PCR detection of CMV in R. solani strains (B) or N. benthamiana plants (E). ssRNA fraction extracted from R. solani strains or N. benthamiana leaves were used for RT-PCR using primer sets specific for CMV RNA 1, 2, 3 and R. solani beta-tubulin. PCR products were run on agarose gel electrophoresis and stained with ethidium bromide (EtBr). (C) dsRNA profiles of R. solani strain infected with CMV or cured. dsRNAs were extracted from total RNAs of fungi, run on polyacrylamide gel electrophoresis, and stained with EtBr. dsRNA extracted from leaves of CMV-infected N. benthamiana plants was included in the gel electrophoresis (Nb/CMV-Fny). (D) Virus symptom expressions in N. benthamiana plants mechanically rub-inoculated with CMV-Fny virions or total RNA extracted from R. solani carrying CMV-Rs. Plants were photographed at 7 dai. CP-sub, CP subgenomic RNA; N.b., N. benthamiana.