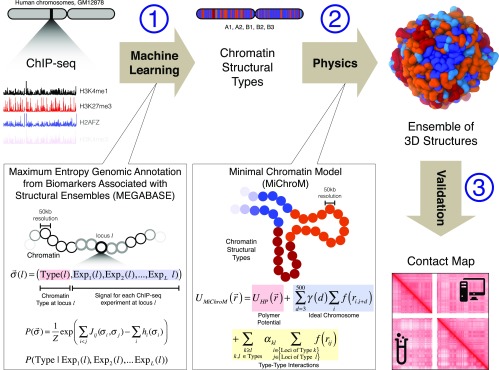
Fig. 1.
Schematic illustration of the MEGABASE + MiChroM computational pipeline. (1) ChIP-Seq data constitute the only input to our pipeline. ChIP-Seq tracks obtained from a publicly available resource (ENCODE) are converted into a sequence of chromatin structural types using a neural network dubbed MEGABASE. The neural network encodes the relationship between compartmentalization and the biochemical state of each locus along the genome. (2) Sequences of chromatin structural types are used as input to a physical model for chromatin folding (MiChroM) to obtain the ensembles of 3D structures of specific chromosomes (17). MiChroM is an effective energy landscape model consisting of a generic polymer with chromatin-type interactions and a translational invariant local ordering term (Ideal Chromosome). (3) Ensembles of 3D structures are validated by comparing the predicted contact maps with those experimentally determined by using Hi-C.