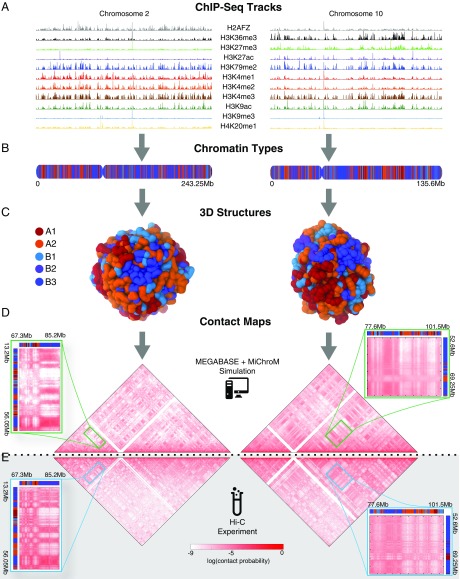
Fig. 2.
Predicting the 1D chromatin sequences, 3D conformations, and 2D contact probabilities of human chromosomes from epigenetic marking patterns. We apply MEGABASE + MiChroM to obtain an ensemble of 3D structures for all of the autosomes of cell line GM12878. For illustrative purposes, predictions for chromosome 2 (Left) and chromosome 10 (Right) are shown, respectively. (A) Ninety-five ChIP-Seq tracks are downloaded from the ENCODE database and used as input for MEGABASE to predict 1D sequences of chromatin types (shown in B). The 3D structure of each chromosome is encoded in its specific 1D sequence of chromatin structural types. (C) Typical 3D conformation obtained by MiChroM is shown for chromosomes 2 and 10. (D) Approximately 50,000 structures are collected from simulation to generate high-quality contact maps. These contact maps are compared with the Hi-C maps shown in E. The simulations correctly predict the long-range contact probability patterns that are observed in Hi-C maps, as seen in the magnified regions.