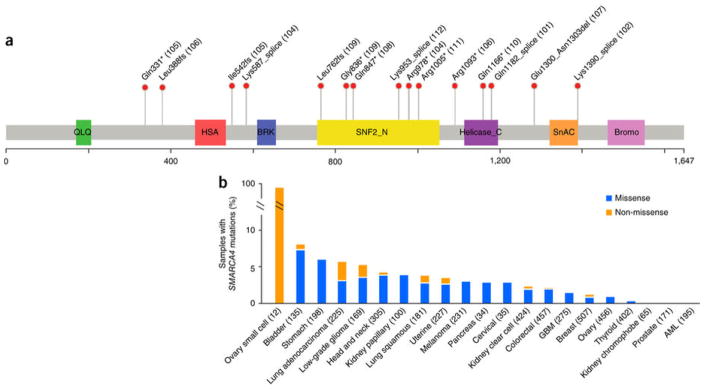
Figure 1. SMARCA4 mutations in SCCOHT and TCGA samples.
(a) Domain structure of the SMARCA4 protein (UniProt, SMCA4_HUMAN) overlaid with the alterations identified in 11 of the 12 SCCOHT cases in this study (case numbers in parentheses; case 103 with exon deletion is not shown). SNF2_N, SNF2 family N-terminal domain; helicase, helicase-conserved C-terminal domain; SnAC, Snf2-ATP coupling, chromatin- remodeling complex; bromo, bromodomain. Genomic coordinates for splice-site mutations can be found in Supplementary Table 5. (b) Percentages of samples with nonsynonymous SMARCA4 mutations in SCCOHT and TCGA non-hypermutated samples (numbers of samples per study in parentheses). Blue bars represent samples with missense-only mutations, and orange bars represent samples with non-missense (including nonsense, frameshift, splice-site and indel) mutations. GBM, glioblastoma multiforme; AML, acute myeloid leukemia.