Abstract
BACKGROUND
External quality assessment (EQA) with commutable samples is essential for assessing the quality of assays performed by laboratories, particularly when the emphasis is on their standardization status and interchangeability of results.
METHODS
We used a panel of 20 fresh-frozen single-donation serum samples to assess assays for the measurement of creatinine, glucose, phosphate, uric acid, total cholesterol, HDL cholesterol, LDL cholesterol, and triglycerides. The commercial random access platforms included: Abbott Architect, Beckman Coulter AU, Ortho Vitros, Roche Cobas, Siemens Advia, and Thermo Scientific Konelab. The assessment was done at the peer group level and by comparison against the all-method trimmed mean or reference method values, where available. The considered quality indicators were intraassay imprecision, combined imprecision (including sample–matrix interference), bias, and total error. Fail/pass decisions were based on limits reflecting state-of-the-art performance, but also limits related to biological variation.
RESULTS
Most assays showed excellent peer performance attributes, except for HDL- and LDL cholesterol. Cases in which individual assays had biases exceeding the used limits were the Siemens Advia creatinine (−4.2%), Ortho Vitros phosphate (8.9%), Beckman Coulter AU triglycerides (5.4%), and Thermo Scientific Konelab uric acid (6.4%), which lead to considerable interassay discrepancies. Additionally, large laboratory effects were observed that caused interlaboratory differences of >30%.
CONCLUSIONS
The design of the EQA study was well suited for monitoring different quality attributes of assays performed in daily laboratory practice. There is a need for improvement, even for simple clinical chemistry analytes. In particular, the interchangeability of results remains jeopardized both by assay standardization issues and individual laboratory effects.
Performing accurate and precise measurements that are comparable over time and location and across assays is essential for ensuring appropriate clinical and public health practice. One step toward achieving this goal is using assays that are metrologically traceable to a higher-order reference measurement system or harmonized by use of internationally recognized procedures (1, 2). In Europe, the European Union Directive on in vitro diagnostic medical devices requires demonstration of metrological traceability (3). Thus, in principle, laboratories using CE-marked assays consisting of calibrator, reagent, and instrument from the same manufacturer (so-called homogeneous systems) can assume accuracy and interchangeability of their measurement results. However, the intrinsic quality of a manufacturer’s assay or test system might be confounded by the laboratory using the system. Therefore, an independent assessment of the quality of measurements obtained under routine conditions is essential for ensuring optimal patient care and public health. External quality assessment (EQA),6 also called proficiency testing, plays a key role in this regard (4, 5). To cover the broad spectrum of sources potentially invalidating the quality of measurements, the assessment schemes need to fulfill certain requirements in terms of design and interpretation (6–9). Early studies described the potentials and limitations of using fresh-frozen single-donation sera for EQA (10–13). Building on findings from these studies, there has been an increasing interest in using serum materials that are commutable, meaning they closely resemble the relevant properties of real patient samples (14–18). However, many EQA schemes continue to use highly processed (mostly lyophilized) and therefore, potentially non-commutable blood products. This practice necessarily limits the scope of these programs to the assessment of laboratory performance at the peer group level.
Here we report on a recent EQA survey that is designed to be not confounded by commutability issues and which enables the assessment of the quality of assays as performed by clinical laboratories. Special emphasis was put on the standardization status of the assays, meaning the accuracy and interchangeability of results across manufacturers and laboratories. We choose assays available on modern random-access test systems for measurement of 8 common analytes, i.e., creatinine, glucose, phosphate, uric acid, total cholesterol, HDL cholesterol, LDL cholesterol, and triglycerides. We carefully selected quality indicators and state-of-the-art limits to reflect both assay and laboratory performance.
Data Analysis
STUDY DESIGN AND SAMPLES
We performed this study with 20 fresh-frozen single-donation serum samples obtained from Solomon Park Research Laboratories. Serum was collected according to the CLSI protocol C37-A without filtration and with 2 U/mL human thrombin (Sigma-Aldrich) added to the serum to facilitate clotting at room temperature (19 ). The individual blood donations were tested and found negative for anti-HIV I/II, anti–hepatitis C virus, and hepatitis B surface antigen. Immediately after 1-mL portions of the sera were aliquotted into polypropylene vials, the sera were stored at −70 °C and kept under these storage conditions until shipment on dry ice to the participating laboratories (see Table 1 in the Data Supplement that accompanies the online version of this report at http://www.clinchem.org/content/vol60/issue6). The samples were required to be kept frozen until analysis. The participants each received 1 aliquot of the 20 samples, which was sufficient for analysis of the 8 analytes in singlet in the same run. The number and selection of participants (63 in total) was adapted to obtain peer groups of almost equal size (see below) and to represent the following manufacturers/test systems: Abbott Architect (n = 10), Beckman Coulter AU (n = 12), Ortho Vitros (n = 11), Roche Cobas (n =10), Siemens Advia (n =12), and Thermo Scientific Konelab (n = 8). Details of the system types, assays, and measurement principles are provided in online Supplemental Table 2. Because of too few (n = 5) laboratories reporting results, no peer group could be established for the Beckman and Ortho LDL cholesterol assays, nor for the Thermo Scientific phosphate assay, and generally not for the Jaffe creatinine assay. However, for LDL cholesterol and phosphate, the respective manufacturers agreed to fill the gap of results by participation with their application laboratories and multiple instruments, so that pooling of the manufacturer/laboratory data was possible. Data assessment was done at the peer group level and by comparing results with target values obtained from either calculating the all-method trimmed mean (AMTM) or, in case of total cholesterol, creatinine, and uric acid, from reference methods (further referred to as REF) (20–22).
STATISTICAL DATA TREATMENT
All numerical results were converted to Système International d’Unités (SI) units. If a laboratory applied a factor to its results when reporting, the results were converted to the originally measured values. Single outlying results reported by a participant were identified and removed by their z-value (>4) based on the median SD for the 20 samples of the respective peer group. Outlying laboratories in a peer group were identified on the basis of the mean of their results for the 20 samples by a 2-sided Grubbs test (95% probability) and omitted from calculation of the peer group mean. Outlying assays were also identified by the Grubbs test (23 ). The AMTM was calculated as mean of the peer group means, after omission of the outlying assays.
QUALITY INDICATORS AND PERFORMANCE LIMITS
The quality indicators applied for performance assessment of both the individual laboratory and assay were (a) intraassay imprecision, (b) combined imprecision (including sample-related effects), (c) bias, and (d) total error (TE). Indicators a, c, and d were estimated at both the peer group and reference level by assessing the data against the peer group mean and AMTM/REF, respectively. Estimation of the combined imprecision (indic.ator b) required comparison of the data with the AMTM/REF, as applicable. The assays were also assessed for peer variation. Estimation of the quality indicators at the peer group level was done as follows. The intraassay imprecision of a laboratory was derived from the Sy|x (expressed as percentage to the mean) from linear regression of its results for the 20 samples in the same run against the peer group mean and referred to as the “laboratory peer Sy|x” (note: the Sy|x is the standard error of estimate or SD of the individual results about the best-fit line). The intraassay imprecision of an assay was estimated from the “median” laboratory peer Sy|x (referred to as “assay median peer Sy|x”). The peer variation of an assay was estimated from the %CV of the laboratory data predicted from linear regression to the peer results at 3 concentrations (low, mid, and high, “peer CV”). The bias and TE for a laboratory were derived, respectively, from the percentage deviation of its results from the peer group targets at 3 concentrations (“laboratory peer bias”) and by the calculation: “laboratory peer TE” = laboratory peer bias + 1.645 × laboratory peer Sy|x; the TE for an assay (“assay peer TE”) was also calculated as 1.96 × square root(peer CV2 + assay median peer Sy|x2). Estimation of the quality indicators at the reference level was done for both the laboratories and the assays as described above, but against the AMTM/REF. The respective estimates were referred to as the “laboratory AMTM/REF Sy|x,” and “assay median AMTM/REF Sy|x” (reflecting, as explained above, the combined imprecision), “laboratory or assay AMTM/REF bias,” and “laboratory or assay AMTM/REF TE.” Calculation of the latter was done with use of the respective laboratory or assay AMTM/REF estimates for bias and Sy|x. The quality indicators were assessed against fixed limits tailored to result in a 5% failure rate. In some cases, the limits were individually expanded to account for either the uncertainty of the respective targets or the inflation of Sy|x due to combined imprecision effects. The assay AMTM/REF bias was additionally assessed against limits derived from the biological variation concept (optimal bias according to the diagnosis model) (24, 25 ). Online Supplemental Tables 3 through 6 provide a full description of the used limits. Online Supplemental Fig. 1 illustrates the estimates of some of the above quality indicators.
Results
OUTLIER IDENTIFICATION
Eighteen single outliers out of 9900 results (0.2%) were identified. For 11 laboratories the results for 1 analyte (out of 495 tests in total, i.e., 8 analytes measured in 63 laboratories minus 9 missing values, or 2.2%) were omitted as outliers from their peer group. The following assays were identified as outliers and omitted from calculation of the AMTM: Ortho Vitros phosphate, Beckman AU triglycerides, and Thermo Scientific Konelab uric acid.
ASSAY PERFORMANCE
Peer estimates
The limits for assay median peer Sy|x (1.5%) were exceeded for 3 assays (1 for creatinine, 2 for HDL cholesterol). The limits for the peer CV (3%) were occasionally violated at the mid and high concentrations (HDL- and LDL cholesterol, creatinine), but more frequently at the low end (in particular creatinine, HDL cholesterol, triglycerides). The peer TE of 3 assays (creatinine, HDL cholesterol, triglycerides) exceeded the 6.5% limit. The “median” of the respective estimates across manufacturers was on the order of 1% (assay median peer Sy|x), 2% (peer CV at mid and high concentrations), 2%–3% (peer CV at low concentration, rising to 5% for triglycerides), and 4%–5% (assay peer TE). Online Supplemental Table 7 provides full numerical documentation.
AMTM/REF estimates
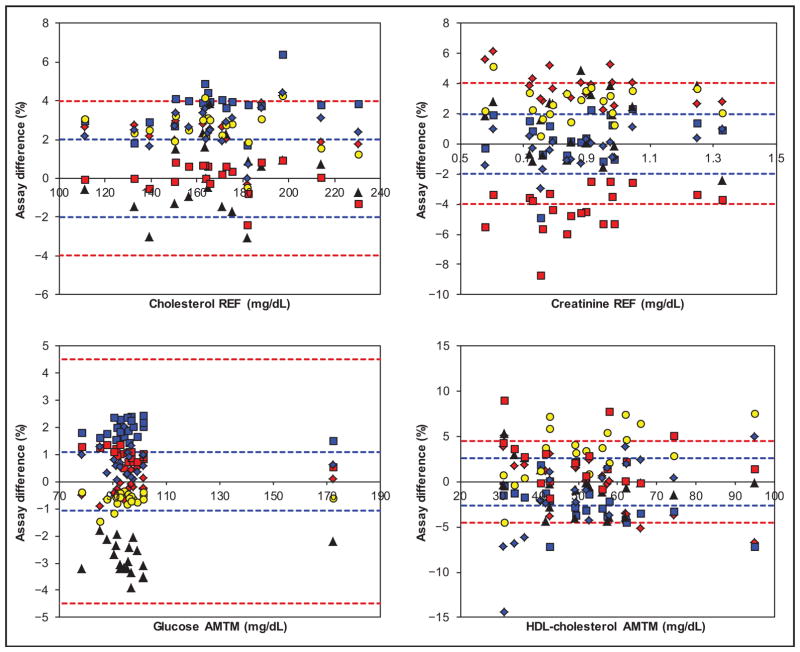
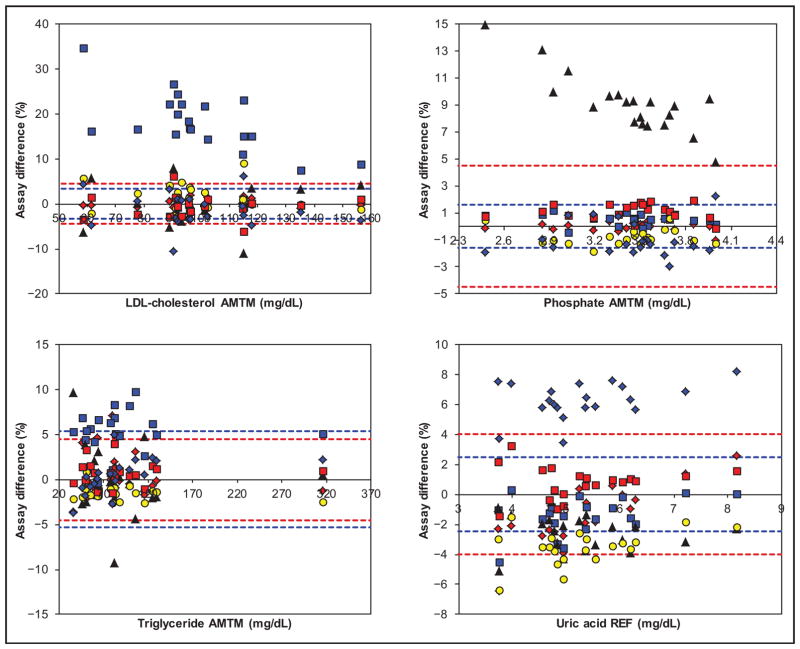
Figs. 1 and 2 show the assay AMTM/REF bias and TE estimates vs analyte-specific fixed and optimal biological bias limits. Online Supplemental Figs. 2 and 3 show the same data with SI units in the x axis. Table 1 documents the assay AMTM/REF bias at low, mid, and high concentrations for each of the manufacturers. For cholesterol, all assays performed within the 4% fixed bias limits (Beckman AU was borderline at high concentration), but not within the 2% optimal limit. For creatinine, the difference between the most biased assays (Abbott Architect and Siemens Advia) was approximately 8%; at the low concentration the discrepancy was even higher because of the tendency of the Abbott assay to an increase in bias. The 2% optimal limit was challenging for several assays. For glucose, the results of all assays showed a narrow distribution. Notably, the optimal limit of approximately 1% was beyond current technical capabilities; concentration-related biases seemed to be absent, because the observed differences at high concentrations fitted well to the rest of the differences. For HDL cholesterol, the difference between the most biased assays (Roche Cobas and Beckman AU) was approximately 7%; the biases depended on the concentration for several assays, which may need confirmation by extensive comparison with an REF). The LDL cholesterol percentage difference plot is dominated by the high and concentration-dependent bias of the Beckman assay (>15%) and the generally large variability of the other assays (the Abbott assay was a notable exception). Note that for the Beckman and Ortho assays the difference plot is based on the pooled manufacturer/laboratory data (differently from the data in Table 1). The comparability of 5 assays was borderline but within the optimal limit of 3.5%. For phosphate, the comparability of the assays (except Ortho) was excellent, with differences even within the optimal limit of 1.6%. In contrast, the Ortho assay had a positive bias (8.9%) that increased at lower concentrations. Note that the Thermo Scientific phosphate results in the difference plot were also pooled manufacturer/laboratory data. For triglycerides, the comparability of the assays (except Beckman) was very good, with differences within the optimal limit of 5.3%; concentration-related effects seemed to be absent. For uric acid, the comparability of the assays (except Thermo Scientific showing a bias of 6.4%) was very good; the optimal limit of 2.5% was challenging and not met. For the Roche Cobas and Abbott Architect, there are indications for increased negative bias at low concentrations. The regression and correlation data (assay peer data against the AMTM/REF) are summarized in online Supplemental Table 8.
Fig. 1. Assay percentage difference for cholesterol, creatinine, glucose, and HDL cholesterol vs AMTM or REF target values, as applicable, for Abbott (red diamond), Beckman (blue square), Ortho (black triangle), Roche (yellow circle), Siemens (red square), and Thermo (blue diamond).
The red-broken bias limits are those listed in online Supplemental Table 5; the blue-broken limits are optimal bias limits from biological variation (see online Supplemental Table 6) (24, 25 ). For conversion of the traditional units to SI units used in the online supplemental figures, multiply by 0.02586 for cholesterol (mmol/L), 88.40 for creatinine (μmol/L), 0.05551 for glucose (mmol/L), and 0.02586 for HDL cholesterol (mmol/L).
Fig. 2. Assay percentage difference for LDL cholesterol, phosphate, triglycerides, and uric acid vs AMTM or REF target values, as applicable, for Abbott (red diamond), Beckman (blue square), Ortho (black triangle), Roche (yellow circle), Siemens (red square), and Thermo (blue diamond).
The red and blue broken limits are the same as described for Fig. 1. For conversion of the traditional units to SI units used in online supplemental figures, multiply by 0.02586 for LDL cholesterol, 0.3229 for phosphate (mmol/L), 0.01129 for triglycerides (mmol/L), and 59.48 for uric acid (μmol/L). due to a favorable combination of substantial bias (−4.2%) but excellent median Sy|x (1.6%).
Table 1.
AMTM/REF bias estimates (at low, mid, and high concentrations) for each assay (named by manufacturer).a
| CHOLb | CREA | GLU | HDL | LDL | PHOS | TRIGL | UA | |
|---|---|---|---|---|---|---|---|---|
| Bias limit (%) | 4 | 4 | 4.5 | 4.5 | 4.5 | 4.5 | 4.5 | 4 |
| Abbott | 3.0 | 5.1 | −0.4 | 5.5 | 2.7 | 0.1 | 2.5 | −4.9 |
| 2.5 | 3.7 | −0.2 | −1.0 | 1.5 | 0.0 | 0.1 | −1.2 | |
| 2.3 | 2.8 | 0.2 | −5.4 | 0.9 | 0.0 | −1.0 | 1.9 | |
| Beckman | 2.7 | −0.6 | 2.1 | 0.3 | NA | 0.8 | 5.8 | −2.6 |
| 3.8 | 0.5 | 1.9 | −3.2 | NA | 0.5 | 5.4 | −1.3 | |
| 4.4 | 1.1 | 1.6 | −5.6 | NA | 0.4 | 5.3 | −0.3 | |
| Ortho | −0.6 | 1.6 | −3.0 | 0.0 | NA | 14.6 | −0.9 | −2.2 |
| 0.0 | 1.2 | −2.8 | −1.1 | NA | 8.9 | −0.3 | −2.6 | |
| 0.4 | 0.9 | −2.4 | −1.9 | NA | 6.7 | 0.0 | −2.8 | |
| Roche | 3.5 | 2.6 | −0.7 | −0.3 | 5.2 | −1.1 | 0.0 | −4.6 |
| 2.5 | 2.7 | −0.6 | 3.9 | 2.0 | −0.7 | −1.6 | −3.4 | |
| 1.9 | 2.7 | −0.6 | 6.7 | 0.4 | −0.5 | −2.3 | −2.4 | |
| Siemens | 0.7 | −5.5 | 1.2 | 2.8 | 0.0 | 1.3 | 0.3 | 0.4 |
| 0.0 | −4.2 | 1.0 | 2.2 | −0.3 | 1.1 | 0.7 | 0.8 | |
| −0.4 | −3.4 | 0.5 | 1.7 | −0.4 | 1.1 | 1.0 | 1.2 | |
| Thermo Scientific | 2.2 | −1.5 | 0.8 | −8.3 | 2.1 | NA | −1.9 | 5.2 |
| 2.6 | −0.3 | 0.7 | −0.7 | −0.1 | NA | 1.0 | 6.4 | |
| 2.8 | 0.5 | 0.6 | 4.4 | −1.3 | NA | 2.3 | 7.3 |
The underlined values indicate violation of the limits.
CHOL, cholesterol; CREA, creatinine; GLU, glucose; HDL, HDL cholesterol; LDL, LDL cholesterol; PHOS, phosphate; TRIGL, triglycerides; UA, uric acid; NA, not applicable.
Online Supplemental Table 9 provides a detailed overview of all AMTM/REF estimates (bias figures reiterated but with CIs) and shows that the “median” of the assay median AMTM/REF Sy|x (%) estimates across manufacturers was remarkably similar to the peer equivalent (see online Supplemental Table 7) for cholesterol, creatinine, glucose, phosphate, and uric acid; however, the “median” was considerably increased for HDL cholesterol (2.6% vs 1%), and LDL cholesterol (2.9% vs 1%), the latter mainly due to the high values for the Roche, Siemens, and Thermo Scientific assays. Note that in online Supplemental Table 9, no values are given for the assay median AMTM/REF Sy|x for triglycerides because of the inconsistency of measurement data. From the tabulated assay AMTM/REF TE estimates, all assays that violated the fixed bias limits also did so for TE, except the Siemens creatinine assay,
LABORATORY PERFORMANCE
Table 2 shows the bias (vs AMTM) observed at 3 concentrations for the 63 participating laboratories. Whereas biases represent the combined effects of laboratory and assay, the interlaboratory differences observed here were particularly influenced by assay bias. Laboratories with maximum absolute biases >15% were regularly observed, frequently in the low concentration range. Consequently, this led to differences of > 30% between the highest deviating laboratories (=Diff 1), and differences of >15% between the third most deviating laboratories (= Diff 3).
Table 2.
Observed AMTM bias in the participating laboratories.a
| CHOLb | CREA | GLU | HDL | LDL | PHOS | TRIGL | UA | |
|---|---|---|---|---|---|---|---|---|
| Bias (%) | ||||||||
| Minimum | −16 | −9 | −8 | −20 | −17 | −6 | −15 | −7 |
| Maximum | 6 | 17 | 6 | 8 | 25 | 13 | 13 | 11 |
| Diff 1c | 22 | 26 | 14 | 27 | 43 | 19 | 28 | 18 |
| Diff 2 | 11 | 22 | 10 | 16 | 25 | 14 | 21 | 16 |
| Diff 3 | 9 | 17 | 9 | 13 | 21 | 14 | 15 | 13 |
| Bias low (%) | ||||||||
| Minimum | −18 | −10 | −10 | −16 | −22 | −8 | −35 | −7 |
| Maximum | 6 | 28 | 6 | 14 | 42 | 21 | 29 | 14 |
| Diff 1 | 24 | 38 | 16 | 30 | 64 | 29 | 64 | 21 |
| Diff 2 | 12 | 36 | 12 | 22 | 52 | 22 | 38 | 18 |
| Diff 3 | 9 | 35 | 10 | 19 | 39 | 21 | 23 | 14 |
| Bias high (%) | ||||||||
| Minimum | −15 | −9 | −6 | −22 | −15 | −6 | −14 | −7 |
| Maximum | 6 | 10 | 6 | 10 | 17 | 10 | 9 | 10 |
| Diff 1 | 21 | 19 | 12 | 32 | 32 | 16 | 23 | 17 |
| Diff 2 | 11 | 17 | 11 | 18 | 16 | 12 | 13 | 14 |
| Diff 3 | 9 | 16 | 8 | 18 | 14 | 11 | 11 | 13 |
The underlined values refer to maximum absolute laboratory biases >15% and differences between laboratories >30%.
CHOL, cholesterol; CREA, creatinine; GLU, glucose; HDL, HDL cholesterol; LDL, LDL cholesterol; PHOS, phosphate; TRIGL, triglycerides; UA, uric acid.
Diff 1, the difference between the most deviating laboratories; Diff 2 and 3, the differences between the second and third most deviating laboratories.
Discussion
We designed our study for adequacy to assess different performance attributes of assays performed on modern platforms and used in a clinical setting. An important design attribute was the collection and processing of the single-donation samples according to the best available protocol to warrant commutability (CLSI C37-A). This allowed us to reliably investigate the comparability of assays across manufacturers. The compelling observations in this regard show the utility of organizing specially designed surveys complementary to those in common EQA schemes. Figs. 1 and 2 show that even for simple clinical chemistry analytes the standardization status of certain assays is still a matter of concern. Three assays had to be excluded from the AMTM because of a statistically significant bias (phosphate, triglycerides, uric acid), and the pooled manufacturer/laboratory data for one LDL cholesterol assay suggested a 15% bias; however, this needs confirmation (see below). In addition, it was striking to find differences between assays up to 8% (creatinine, HDL cholesterol), and frequent concentration-related biases (particularly for HDL cholesterol). Altogether, these observations point to considerable calibration differences between manufacturers/assays. The data further show that assays do not yet meet the optimal bias limits necessary for clinical use (24 ). Particularly striking in this regard was that this also applied for cholesterol and creatinine, in spite of dedicated standardization programs such as the National Cholesterol Education and National Kidney Disease Education Program. A recent impact study from the College of American Pathologists reported similar across-assay differences for creatinine (26 ). In contrast, the biological bias limits were met for phosphate, LDL cholesterol, triglycerides, and uric acid, except for the aforementioned biased assays (Ortho phosphate, Beckman triglycerides and LDL cholesterol, and Thermo Scientific uric acid).
Whereas the more advanced classical EQA surveys, i.e., those using pooled samples obtained with adherence to the C37-A recommendations, may also adequately reveal standardization issues, our design with 20 single-donation samples had the additional potential to assess the intraassay imprecision, combined imprecision, and TE. Indeed, pooling potentially dilutes matrix interferences present in individual samples. The assay peer performance estimates (see online Supplemental Table 7) illustrated excellent intraassay imprecision (often <1.5%), and mostly low variation (<3%) at mid and high concentrations, but significantly increased at the low end, and a TE in most cases of <6.5%, except for 3 assays. The high dispersion of some data for triglycerides may be related to glycerol contamination. Assessment of assay performance against the AMTM/REF showed good analytical specificity for cholesterol, creatinine, glucose, phosphate, and uric acid (“median” AMTM/REF Sy|x across manufacturers remarkably similar to the peer equivalent, in spite of individual differences). In contrast, the considerably increased values for this estimate for all HDL cholesterol and LDL cholesterol assays may point to sample-related effects, as shown elsewhere (27 ). In general, our findings demonstrate that assays for HDL-and LDL cholesterol have not yet reached the quality present for the other analytes, although we must admit that the accuracy of assays for these analytes may be affected by preanalytical and other conditions (e.g., dyslipidemia, diabetes). In this regard, the manufacturer of the LDL cholesterol assay with the bias of >15% argued that it has been shown that sample freezing potentially confounds performance (28 ). On the other hand, we found in the concerned manufacturer package insert that, if specimens need to be stored for >5 days, they may be preserved at temperatures lower than −70 °C for up to 3 months (29 ). With regard to the assay AMTM/REF TE estimate, we found that, in general, this performance attribute followed the assay AMTM/REF bias.
Our study also reflected on the robustness of the assays’ intrinsic quality for satisfactory performance in a daily laboratory context. Assessment of the laboratory peer group performance showed that 2.1% of the laboratory tests could not be included in the calculation of peer group targets. Laboratory AMTM performance sometimes showed a large difference between laboratories for all estimates. Particularly striking were the differences of >30% between laboratories (Table 2). Although these differences were partly due to biases in the used assays, the existence of laboratory effects was obvious. The regularly to frequently observed maximum absolute biases (>15% and up to 30%) were per se amazing in view of the fact that only 63 laboratories participated in the study.
Our study had limitations. The small amount of sample volume (maximum 180 mL) restricted the number of participants. Nevertheless, reliable statistics could be obtained with only 63 laboratories, provided there was careful selection on the basis of the use of homogeneous test systems. Controlled peer grouping is important to estimate biases because the assays are more influential than the measurement principles (9 ). We also assumed commutability of our samples. However, there is no better alternative than the C37-A protocol to collect native samples in big volumes. This approach has been used in other dedicated EQA schemes, and a review addressing this issue in detail concluded that C37 sera are commutable for many analytes (15, 30, 31 ). Our study may further be questioned with regard to the target setting for bias/traceability assessment, which was not necessarily trueness based (only for 3 of the 8 analytes) (20–22). Some participants deemed our REF targets for cholesterol set with a GC-MS REF inappropriate because their assays were calibrated to the Abell–Kendall method, which has been shown to deviate from GC-MS (32 ). Although this could explain some of the positive biases, we still believe that our observations point to a general lack of standardization of lipid assays. With regard to using the AMTM instead of REF, our study gave conclusive evidence to support that AMTM is a reasonable and cost-saving alternative, at least for the analytes we examined here. Indeed, the AMTMs for cholesterol, creatinine, and uric acid were very similar to the REF values (largest difference only 1.9%) and had an almost identical uncertainty (CIs typically 1.5%) (33, 34 ). Of course, to use the AMTM for assessment of the comparability across assays, its stability over time is required. This might be problematic, given potential variability among assay calibrator and reagent lots. However, reference laboratories also need to have adequate means in place to assure accuracy over time (35, 36 ). All considerations taken together, in future surveys we will set REF targets in the case of inconclusive AMTMs.
Another limitation is that the concentrations covered by samples from apparently healthy volunteers were restricted to the reference range, and consequently the performance at concentrations typically observed in diseased patients could be different from what was observed here. We tried to cope with this limitation to a certain extent by also looking into the quality at the low and high concentrations. In fact, this revealed that sometimes the calibration was only successful in the mid-concentration range. The appropriateness of our limits may also be questioned. We considered the fixed limits used to be state-of-the-art for assessing the performance of modern instruments because they were tailored to a 5% failure rate. Furthermore, for assessing the assay and laboratory performance against the peer mean and AMTM/REF, we expanded the limits analyte specifically to account for the uncertainty of the targets. Finally, we believe that biological bias limits make sense in view of assessing the utility of the assays for clinical purposes (24 ).
In conclusion, our study provided reliable information on assay and laboratory performance in daily practice, with special emphasis on the interchangeability of results across manufacturers and laboratories. Our results are comforting in that the quality of within-peer performance was excellent, but with room for improvement at the higher and lower concentrations. There were some sample-related effects, with isolated cases of deviating assays, differences across manufacturers and laboratories, and the inability to meet the biological bias limits for numerous analytes. On the basis of these observations, we advocate the need to address quality issues in a more targeted manner, e.g., by adaptation of regulatory and EQA practices, but also by educating individual laboratories toward more emphasis on quality assurance. Special studies like ours do not substitute for but are complementary to more common EQA surveys.
Supplementary Material
Acknowledgments
The authors are extremely indebted to the laboratories that subscribed to this study (coordinates in the online supplemental file). Their interest in the study design and objectives, as well as their timely reporting of results, is appreciated. They also were pleased to receive the interest from the different in vitro diagnostic manufacturers.
Footnotes
Nonstandard abbreviations: EQA, external quality assessment; AMTM, all-method trimmed mean; REF, reference methods; SI, Systéme International d’Unités; TE, total error.
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures or Potential Conflicts of Interest: No authors declared any potential conflicts of interest.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
References
- 1.Vesper HW, Thienpont LM. Traceability in laboratory medicine. Clin Chem. 2009;55:1067–75. doi: 10.1373/clinchem.2008.107052. [DOI] [PubMed] [Google Scholar]
- 2.Miller WG, Myers GL, Gantzer ML, Kahn SE, Schönbrunner ER, Thienpont LM, et al. Roadmap for harmonization of clinical laboratory measurement procedures. Clin Chem. 2011;57:1108–17. doi: 10.1373/clinchem.2011.164012. [DOI] [PubMed] [Google Scholar]
- 3.Directive 98/79/EC of the European Parliaments and of the Council of 27 October 1998 on in vitro diagnostic medical devices L331. Off J Eur Communities. 1998;41:1–37. [Google Scholar]
- 4.Sciacovelli L, Secchiero S, Zardo L, Zaninotto M, Plebani M. External quality assessment: an effective tool for clinical governance in laboratory medicine. Clin Chem Lab Med. 2006;44:740–9. doi: 10.1515/CCLM.2006.133. [DOI] [PubMed] [Google Scholar]
- 5.Miller WG. The role of proficiency testing in achieving standardization and harmonization between laboratories. Clin Biochem. 2009;42:232–5. doi: 10.1016/j.clinbiochem.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Stöckl D, Thienpont LM. The combined target approach—a way out of the proficiency testing dilemma. Arch Pathol Lab Med. 1994;118:775–6. [PubMed] [Google Scholar]
- 7.Libeer JC, Baadenhuijsen H, Fraser CG, Petersen PH, Ricós C, Stöckl D, Thienpont L. Characterization and classification of external quality assessment schemes (EQA) according to objectives such as evaluation of method and participant bias and standard deviation. External Quality Assessment (EQA) Working Group A on Analytical Goals in Laboratory Medicine. Eur J Clin Chem Clin Biochem. 1996;34:665–78. [PubMed] [Google Scholar]
- 8.Middle JG, Libeer JC, Malakhov V, Penttilä I. Characterization and evaluation of external quality assessment scheme serum. Discussion paper from the European External Quality Assessment (EQA) Organisers Working Group C. Clin Chem Lab Med. 1998;36:119–30. doi: 10.1515/CCLM.1998.023. [DOI] [PubMed] [Google Scholar]
- 9.Miller WG, Jones GR, Horowitz GL, Weykamp C. Proficiency testing/external quality assessment: current challenges and future directions. Clin Chem. 2011;57:1670–80. doi: 10.1373/clinchem.2011.168641. [DOI] [PubMed] [Google Scholar]
- 10.Stöckl D, Libeer JC, Reinauer H, Thienpont LM, De Leenheer AP. Accuracy-based assessment of proficiency testing results using serum from single donations: possibilities and limitations. Clin Chem. 1996;42:469–70. [PubMed] [Google Scholar]
- 11.Linko S, Himberg JJ, Thienpont L, Stöckl D, De Leenheer A. Assessment of the state-of-the-art trueness and precision of serum total-calcium and glucose measurements in Finnish laboratories—the QSL-Finland study. Scand J Clin Lab Invest. 1998;58:229–39. doi: 10.1080/00365519850186625. [DOI] [PubMed] [Google Scholar]
- 12.Thienpont LM, Stöckl D, Kratochvíla J, Friedecký B, Budina M. Pilot external quality assessment survey for post-market vigilance of in vitro diagnostic medical devices and investigation of trueness of participants’ results. Clin Chem Lab Med. 2003;41:183–6. doi: 10.1515/CCLM.2003.030. [DOI] [PubMed] [Google Scholar]
- 13.Thienpont LM, Stöckl D, Friedecký B, Kratochvíla J, Budina M. Trueness verification in European external quality assessment schemes: time to care about the quality of the samples. Scand J Clin Lab Invest. 2003;63:195–202. doi: 10.1080/00365510310000349. [DOI] [PubMed] [Google Scholar]
- 14.Miller WG, Myers GL, Ashwood ER, Killeen AA, Wang E, Ehlers GW, et al. State of the art in trueness and interlaboratory harmonization for 10 analytes in general clinical chemistry. Arch Pathol Lab Med. 2008;132:838–46. doi: 10.5858/2008-132-838-SOTAIT. [DOI] [PubMed] [Google Scholar]
- 15.Cobbaert C, Weykamp C, Franck P, de Jonge R, Kuypers A, Steigstra H, et al. Systematic monitoring of standardization and harmonization status with commutable EQA-samples–five year experience from the Netherlands. Clin Chim Acta. 2012;414:234–40. doi: 10.1016/j.cca.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 16.Stepman HC, Stöckl D, Acheme R, Sesini S, Mazziotta D, Thienpont LM. Status of serum-calcium and -albumin measurement in Argentina assessed in 300 representative laboratories with 20 fresh frozen single donation sera. Clin Chem Lab Med. 2011;49:1829–36. doi: 10.1515/CCLM.2011.681. [DOI] [PubMed] [Google Scholar]
- 17.Van Houcke SK, Rustad P, Stepman HC, Kristensen GB, Stöckl D, Røraas TH, et al. Calcium, magnesium, albumin, and total protein measurement in serum as assessed with 20 fresh-frozen single-donation sera. Clin Chem. 2012;58:1597–9. doi: 10.1373/clinchem.2012.189670. [DOI] [PubMed] [Google Scholar]
- 18.Stavelin A, Petersen PH, Sølvik UØ, Sandberg S. External quality assessment of point-of-care methods: model for combined assessment of method bias and single-participant performance by the use of native patient samples and non-commutable control materials. Clin Chem. 2013;59:363–71. doi: 10.1373/clinchem.2012.191957. [DOI] [PubMed] [Google Scholar]
- 19.CLSI. CLSI document C37-A. Wayne (PA): CLSI; 1999. Preparation and validation of commutable frozen human serum pools as secondary reference materials for cholesterol measurement procedures; approved guideline. [Google Scholar]
- 20.Edwards SH, Kimberly MM, Pyatt SD, Stribling SL, Dobbin KD, Myers GL. Proposed serum cholesterol reference measurement procedure by gas chromatography-isotope dilution mass spectrometry. Clin Chem. 2011;57:614–22. doi: 10.1373/clinchem.2010.158766. [DOI] [PubMed] [Google Scholar]
- 21.Stöckl D, Reinauer H. Candidate reference methods for determining target values for cholesterol, creatinine, uric acid, and glucose in external quality assessment and internal accuracy control. I. Method setup. Clin Chem. 1993;39:993–1000. [PubMed] [Google Scholar]
- 22.Thienpont LM, De Leenheer AP, Stöckl D, Reinauer H. Candidate reference methods for determining target values for cholesterol, creatinine, uric acid, and glucose in external quality assessment and internal accuracy control. II. Method transfer. Clin Chem. 1993;39:1001–6. [PubMed] [Google Scholar]
- 23.Grubbs F. Procedures for detecting outlying observations in samples. Technometrics. 1969;11:1–21. [Google Scholar]
- 24.Ricos C, et al. Biological variation: from principles to practice. Washington (DC): AACC Press; 2001. [Google Scholar]
- 25.Desirable specifications for total error, imprecision, and bias, derived from intra- and inter-individual biologic variation. [Accessed May 2014]; http://www.westgard.com/biodatabase1.html.
- 26.Killeen AA, Ashwood ER, Ventura CB, Styer P. Recent trends in performance and current state of creatinine assays. Arch Pathol Lab Med. 2013;137:496–502. doi: 10.5858/arpa.2012-0134-CP. [DOI] [PubMed] [Google Scholar]
- 27.Miller WG, Myers GL, Sakurabayashi I, Bachmann LM, Caudill SP, Dziekonski A, et al. Seven direct methods for measuring HDL and LDL cholesterol compared with ultracentrifugation reference measurement procedures. Clin Chem. 2010;56:977–86. doi: 10.1373/clinchem.2009.142810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vesper HW, Wilson PW, Rifai N. A message from the laboratory community to the National Cholesterol Education Program Adult Treatment Panel IV. Clin Chem. 2012;58:523–7. doi: 10.1373/clinchem.2011.178202. [DOI] [PubMed] [Google Scholar]
- 29.LDL-Cholesterol: OSR6196, OSR6296. Brea (CA): Beckman Coulter; [Accessed May 2014]. https://www.beckmancoulter.com/wsrportal/techdocs?docname=/cis/baosr6x96/%25%25/en_ldl-cholesterol.pdf. Package insert information: BAOSR6x96.01, OSR General Chemistry, 2009-08. [Google Scholar]
- 30.Jansen R, Jassam N, Thomas A, Perich C, Fernandez-Calle P, Faria AP, et al. A category 1 EQA scheme for comparison of laboratory performance and method performance: an international pilot study in the framework of the Calibration 2000 project. Clin Chim Acta. 2013 doi: 10.1016/j.cca.2013.11.003. pii:S0009-8981(13)00441-5 [Epub ahead of print 2013 Nov 14] [DOI] [PubMed] [Google Scholar]
- 31.Miller WG. Specimen materials, target values and commutability for external quality assessment (proficiency testing) schemes. Clin Chim Acta. 2003;327:25–37. doi: 10.1016/s0009-8981(02)00370-4. [DOI] [PubMed] [Google Scholar]
- 32.Bernert JT, Jr, Akins JR, Cooper GR, Poulose AK, Myers GL, Sampson EJ. Factors influencing the accuracy of the national reference system total cholesterol reference method. Clin Chem. 1991;37:2053–61. [PubMed] [Google Scholar]
- 33.Thienpont L, Franzini C, Kratochvila J, Middle J, Ricós C, Siekmann L, Stöckl D. Analytical quality specifications for reference methods and operating specifications for networks of reference laboratories. Discussion paper from the members of the external quality assessment (EQA) Working Group B1 on target values in EQAS. Eur J Clin Chem Clin Biochem. 1995;33:949–57. [PubMed] [Google Scholar]
- 34.Stöckl D, Sluss PM, Thienpont LM. Specifications for trueness and precision of a reference measurement system for serum/plasma 25-hydroxyvitamin D analysis. Clin Chim Acta. 2009;408:8–13. doi: 10.1016/j.cca.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 35.Joint Committee for Traceability in Laboratory Medicine (JCTLM) [Accessed January 2014];Database of higher-order reference materials, measurement methods/procedures and services. http://www.bipm.org/jctlm/
- 36.Kessler A, Siekmann L, Weykamp C, Geilenkeuser WJ, Dreazen O, Middle J, Schumann G IFCC Committee on Traceability in Laboratory Medicine (C-TLM) External Quality Assessment Scheme for reference laboratories—review of 8 years’ experience. Clin Chem Lab Med. 2013;51:997–1005. doi: 10.1515/cclm-2012-0722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.