Figure 3.
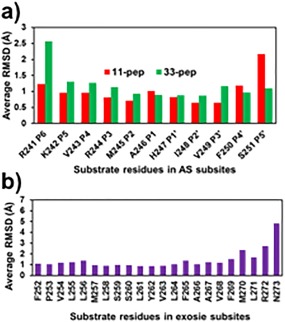
Average per residue RMSD values of substrate backbone atoms (Cα, N, and C). They were calculated by AMBER14 CPPTRAJ algorithm on production trajectory of MD simulated pre‐catalytic non‐covalent complexes formed by GlpG with the substrates. Reference to cluster centroids of conformations relevant for the non‐covalent complex to TC transformation. (A): AS residues of 11‐pep at 350–750 ns, 40,000 frames, and of 33‐pep at 200–450 ns, 25,000 frames. (B): Exosite residues of 33‐pep at 200–450 ns, 25,000 frames.
