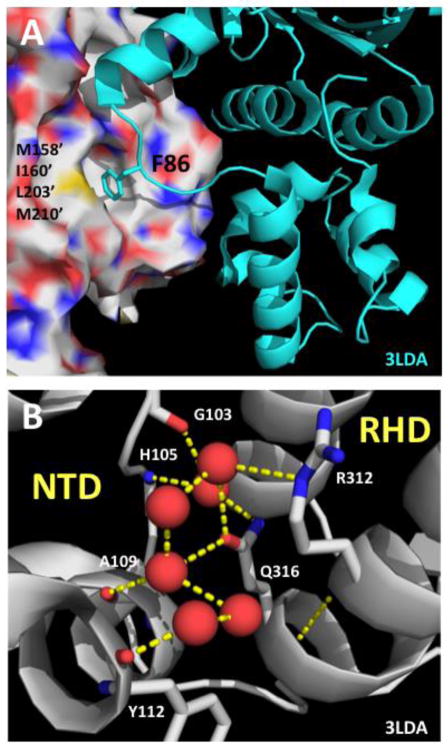
Figure 3. Structural contexts of the F86L and E258A mutations in the RAD51 filament.
(A) Structural context of human RAD51 Phe-86 residue inferred by alignment to a model of the protomer-protomer interface region in the yeast Rad51 filament (PDB ID no. 3LDA). Human RAD51 residue numbering is indicated. Phe-86 from one protomer (cartoon, cyan) binds to a hydrophobic pocket on the adjacent protomer (surface) comprised of residues Met-158’, Ile-160’, Leu-203’, and Met-210’. All of these residues are conserved between human and yeast, except for Met-158’ which is replaced by Leu-216’ in yeast Rad51. (B) Structural context of human RAD51 Glu-258 residue is informed by the structure of the NTD/RHD interface within one protomer of the yeast Rad51 filament (PDB ID no. 3LDA). Yeast Rad51 residue numbering is indicated. Red spheres represent ordered water molecules. A water-mediated hydrogen bond network connects side chains of Gln-316 (equivalent of Glu-258 in human RAD51) and Arg-312 from the RHD to main chain CO and NH functionalities in the NTD.