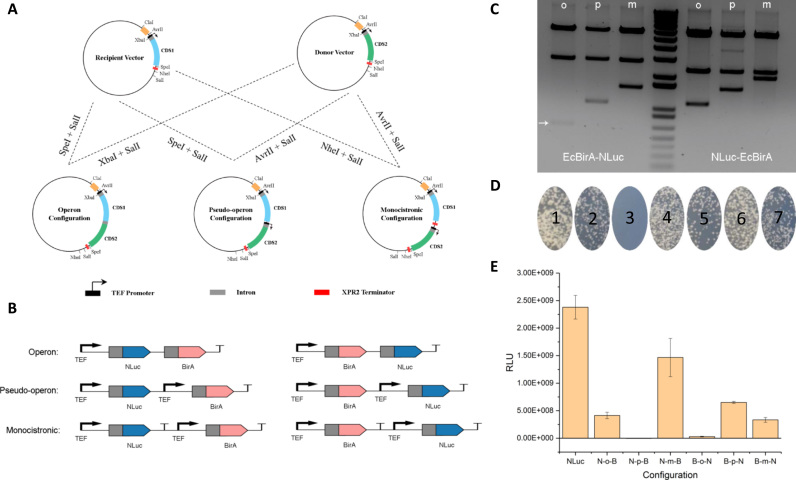
Fig. 3.
Intron alternative splicing allows the construction of three gene configurations in Y. lipolytica. (A) Modular cloning procedure to generate three gene configurations in Y. lipolytica. (B) A detailed layout of a two-gene pathway varying in gene configuration and positional order. (C) Gel digestion pattern of the six gene configurations: lane 1, B-o-N; lane 2, B-p-N, lane 3, B-m-N; lane 4, ladder; lane 5, N-o-B; lane 6, N-p-B and lane 7, N-m-B. (D) Yeast transformation assay to evaluate the efficiency of the Ura3 marker restoring Ura3 auxotrophic phenotype. Plate 1, Ura3 marker positive control; plate 2, U-o-M; plate 3, U-p-M; plate 4, U-m-M; plate 5, M-o-U; plate 6, M-p-U; plate 7, M-m-U. U is an abbreviation for Ura3 and M is an abbreviation for mCherry. (E) Positional effects of gene configuration on gene expression efficiency as evaluated by luminescence readout in the six genetic variations. RLU: relative luminescence unit. Error bars are the standard deviation calculated from triplicate readouts.