Figure 1.
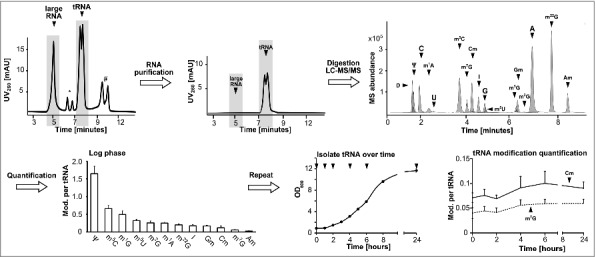
Workflow to determine modification levels in tRNA during yeast growth cycle. The isolated total RNA was purified by size exclusion chromatography (SEC) (*5.8S/5S rRNA peak, #unknown contamination). The tRNA fraction was then digested and the obtained nucleosides separated and quantified over LC-MS to receive the shown chromatogram on the upper right side. The average amounts of the respective modifications per molecule tRNA in log growing cells were calculated and plotted (error bars represent SD of n = 3). The quantification was repeated during growth (arrows indicate sample time points) and the quantities of the modifications Cm and m7G were plotted over time. Abbreviations: D = dihydrouridine, Ψ = pseudouridine, C = cytidine, m1A = 1-methyladenosine, U = uridine, m5C = 5-methylcytidine, m7G = 7-methylguanosine, Cm = 2′-O-methylcytidine, I = Inosine, G = guanosine, m5U = 5-methyluridine, m1G = 1-methylguanosine, Gm = 2′-O-methylguanosine, m2G = N2-methylguanosine, A = adenosine, m22G = N2,N2-dimethylguanosine, Am = 2′-O-methyladenosine.
