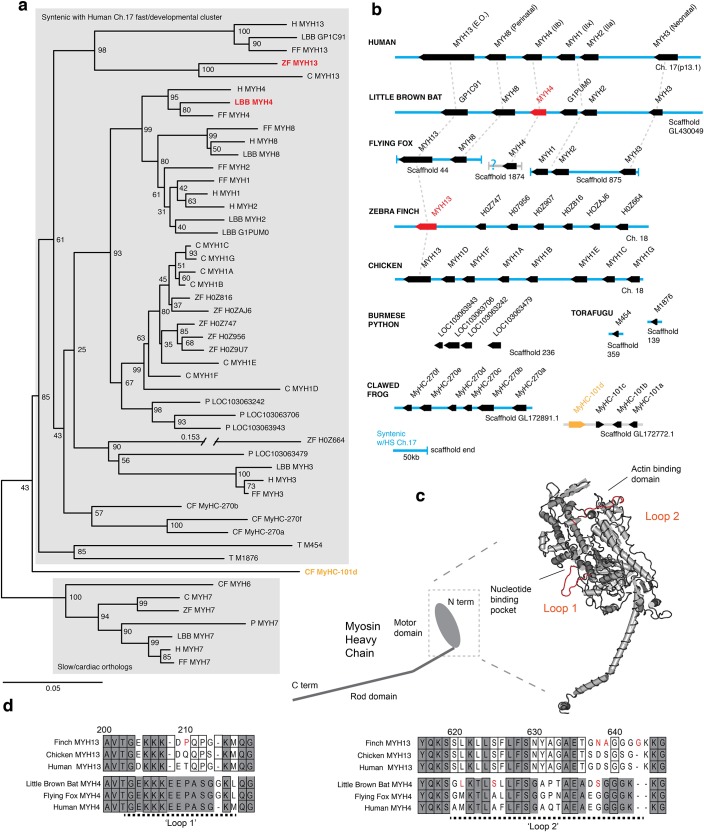
Figure 2. Mammals and avians possess orthologous clusters of myosin heavy chain genes.
(a) Neighbor-joining tree of chicken (C), clawed frog (CF), flying fox (FF), human (H), little brown bat (LBB), Burmese python (P), torafugu (T), and zebra finch (ZF) predicted myosin heavy chain rod domain amino acid sequence from genomic regions syntenic with the human fast/developmental cluster on chromosome 17. Also included are representative cardiac genes, as well as a fast laryngeal myosin gene from the clawed frog (yellow). MYH expressed in bat and finch SFM indicated in red. Human non-muscle MYH9 was used to root the tree (not shown). Branch lengths shown are derived from a maximum likelihood analysis of aligned amino acid sequence. Bootstrap values from a 1000-replicate analysis are given at nodes in percentages. (b) Relative local positions of predicted MYH genes in genomes of the above species. Synteny with human Ch17 is shown in blue. Red and yellow color-coding as in (a). Orthologs indicated by vertical dotted lines. (c) Homology model of human MYH3 myosin motor domain indicating position of loop subdomains and the nucleotide binding pocket. (d) Hypervariable surface loops of MYH13 and MYH4 motor-domains that likely influence actomyosin crossbridge kinetics. Grey shading indicates conservation with/among MYH4 orthologs. Outlines indicate conservation with/among MYH13 orthologs. Substitutions and insertions unique to SFM species in red. Horizontal black dashed lines indicate ‘Loop 1’ and ‘Loop 2’ subdomains.