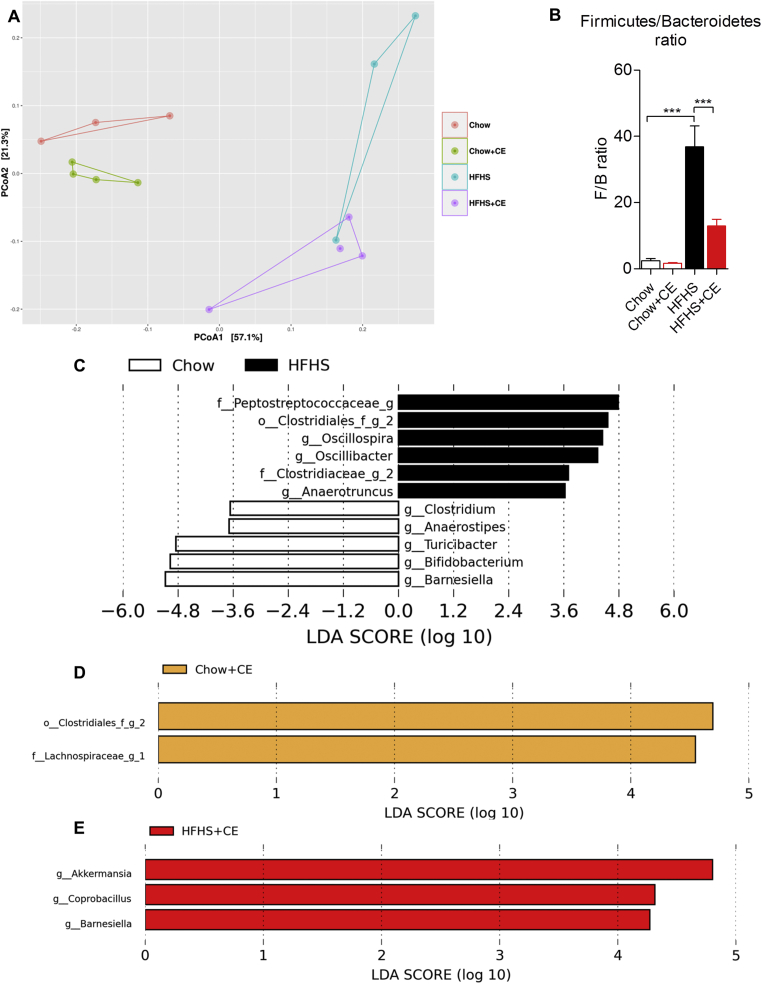
Figure 4.
CE administration alters the taxonomic profile of Chow- and HFHS-fed mice. Genomic DNA was extracted from feces collected at week 21 and subsequent 16S rRNA-based gut microbial profiling was performed. Feces from mice housed in the same cage were pooled and considered as one biological sample (Chow n = 3; Chow + CE n = 4; HFHS n = 3 and HFHS + CE n = 4). (A) β-diversity among groups was initially observed by means of principal component analysis (PCoA) on weighted unifrac distances, and the (B) Firmicutes to Bacteroidetes ratio was calculated as a general index of obesity-driven dysbiosis. Linear discriminant analysis (LDA) effect size (LEfSe) was calculated in order to explore the taxa that more strongly discriminate between the gut microbiota of (C) Chow vs. HFHS, (D) Chow vs Chow + CE and (E) HFHS vs HFHS + CE. (B) Two-way ANOVA with a Student-Newman-Keuls post hoc test was applied to calculate the significance of the differences between groups. *P < 0.05, **P < 0.01 and ***P < 0.001.