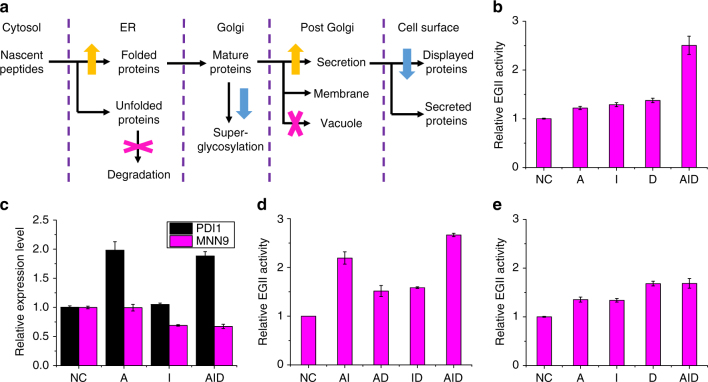
Fig. 4.
CRISPR-AID for combinatorial metabolic engineering. a Yeast surface display of recombinant proteins as a representative example of combinatorial metabolic engineering. Protein folding and secretory machinery, protein super-glycosylation and other surface-displayed proteins, and degradation pathways were chosen as the targets for CRISPRa, CRISPRi, and CRISPRd, respectively. b Combinatorial optimization of EGII display on the yeast surface. EGII activities of the FACS enriched optimal combination (AID-FACS16) and those with the corresponding single component (A-pSg221, I-pSg230, and D-pSg205) were measured. c Verification of CRISPRa (PDI1) and CRISPRi (MNN9) for transcriptional regulation using qPCR. d Exploration of the synergistic interactions among activated (PDI1), interfered (MNN9), and deleted (PMR1) metabolic engineering targets. EGII activities of the double mutants, including AI-pSg417, AD-pSg418, and ID-pSg419, were measured. e Single-factor optimization of EGII display on the yeast surface. EGII activities of the strains with one gRNA (A-pSg218, I-pSg204, and D-pSg186) and the combination of the ones with the highest activities in each category (AID-pSg257) were measured. Error bars represent the mean ± s.d. of biological triplicates