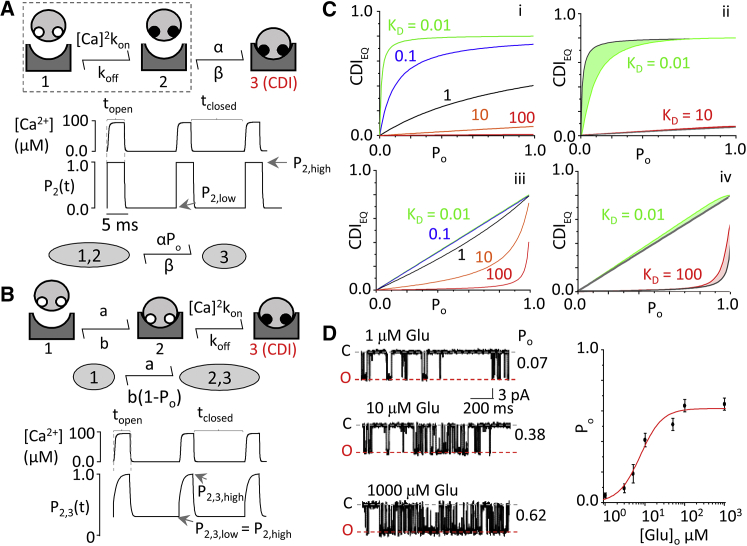
Figure 5.
CDI models reveal unique, testable behaviors. (A) (Top) Model 1 represents NMDA receptor association with holoCaM with subsequent Ca2+. apoCaM (open white circles) binds Ca2+ (solid circles) before binding to the channel. (Middle) Time-dependent behavior of subsystem (dashed box) is given, representing Ca2+ binding to CaM using only fast, N-lobe kinetics in response to a train of Ca2+ influx from a gating channel. State 2 (CaM bound with Ca2+) of the subsystem is pulsatile between P2,high and P2,low occupancy probabilities, in tight synchrony with gating. (Bottom) Shown is simplification of a three-state model given rapid equilibration of its subsystem. (B) (Top) Model 2 represents NMDA receptor association with apoCaM with subsequent Ca binding. (Middle) Given is a simplified model 2 assuming N-lobe kinetics. (Bottom) Shown is behavior of condensed state 2, 3 in response to a train of Ca2+ in-flux. The pulsatile system oscillates between P2,3,high and P2,3,low. P2,3,low is set by the fraction of channels preassociated with apoCaM at rest, P2,high. (C) (i) Given here is evaluation of Eq. 7’s corresponding model 1 across all values of Po. As CaM affinity for the channel is increased, the system exhibits saturation kinetics. As affinity decreases, the system becomes linear. (ii) Deviations of the simplified model in Eq. 7 from numerical integration to the full three-state model did not affect the overall shape or predicted behavior. (iii) Given here is evaluation of Eq. 8’s corresponding model 2 across all values of Po. As CaM affinity for the channel is increased, the system exhibits a linear trend. As affinity decreases, the system uniquely develops an upward curvature. (iv) Deviations of the simplified model in Eq. 8 from numerical integration in the full three-state model were minimal and did not affect the overall shape or predicted behavior. (D) (Left) Exampled here is single-channel activity elicited by indicated glutamate concentration. (Right) Given here is a calibration curve for glutamate concentration and Po determined from cell-attached single-channel recordings. To see this figure in color, go online.