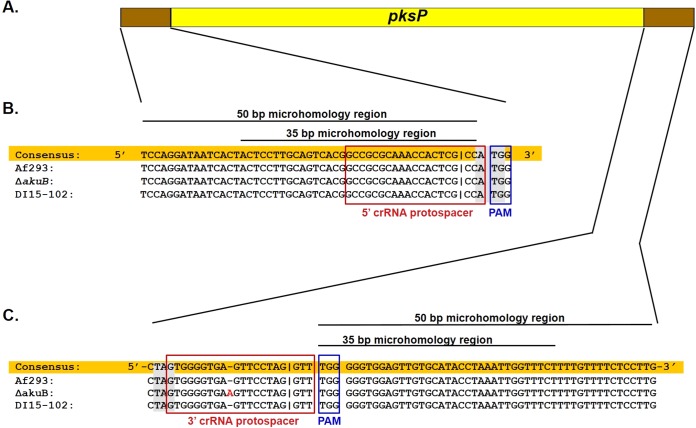
FIG 2 .
Selection of dual crRNA protospacer sequences. (A) Schematic diagram of pksP coding sequence and the flanking regions that are targeted by dual in vitro-assembled Cas9 RNPs. Designing the protospacers is described in Results. (B and C) Sequence alignment of pksP upstream (B) and downstream (C) regions of three distinct genetic backgrounds of A. fumigatus. The consensus sequence (highlighted in orange) was manually generated based on sequence alignment. The 5′ crRNA and 3′ crRNA protospacer sequences are marked by red open boxes. The protospacer-adjacent motif (PAM) sequences are marked by blue open boxes. Start and stop codons are highlighted in gray. The additional adenine in the pksP downstream region of the ΔakuB strain and the clinical isolate DI15-102 is shown in red font. Cas9 DSB sites (i.e., 3 nucleotides upstream of the PAM site [1, 9, 57]) are marked by a vertical line in the sequence. The sequences of the 35-bp and 50-bp regions that are used for microhomology-mediated integration are marked by a line above the sequence.