Fig. 1.
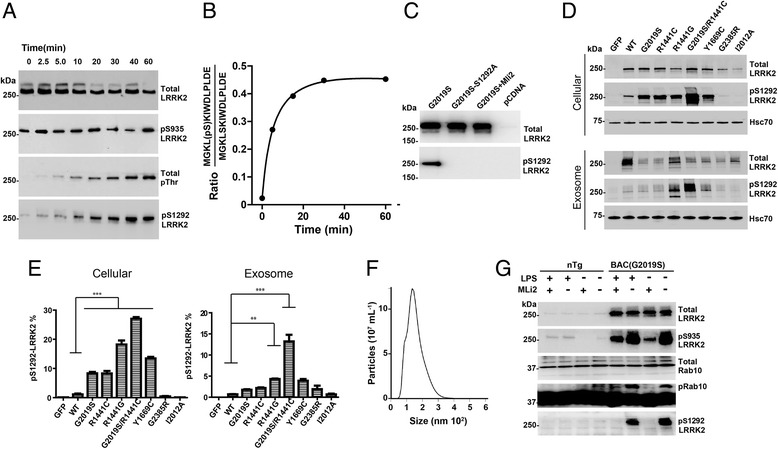
Generation of recombinant pS1292-LRRK2 standards and correlation of exosome and cellular pS1292-LRRK2 levels. a Recombinant G2019S-LRRK2 protein was incubated in kinase reaction buffer for the indicated time. Representative immunoblots are shown. b LC-MS analysis of the pS1292 peptide isolated from recombinant protein from panel A. c Representative immunoblots of S1292A-LRRK2 protein, expressed in HEK293 cells, to validate the authenticity of pS1292-LRRK2 signal. d Representative immunoblots and quantification of HEK293 cell and exosomal lysates, 48-hours after transfection with the indicated LRRK2-encoding plasmids. eGFP-only controls, WT-LRRK2, pathogenic mutations G2019S, R1441C/G, Y1699C, and the double-LRRK2 mutation G2019S/1441C are indicated. Exosome-depleted cell media were incubated with the transfected cells for 24-hours prior to isolation. e Calculated pS1292-LRRK2 levels for cellular and exosome fractions of total LRRK2 protein. Column graphs show mean values with S.E.M. as error bars representing three independent experiments. ***p-value<0.001, **p-value<0.01, *p-value<0.05, ns: p-value>0.05. p value between groups were calculated using Tukey’s multiple comparison test. f Representative nanoparticle tracking analysis of the exosomes isolated from the HEK293 cell cultures in panel C. g Bone marrow derived macrophages from either non-transgenic (Ntg) or G2019S-LRRK2 transgenic mice were treated with LPS to increase pS935-LRRK2 levels, or the LRRK2 kinase inhibitor MLi2 (100 nM) to decrease phospho-LRRK2 levels, as indicated. Rab10 phosphorylation was assessed via phos-tag analysis as described [26]
