Abstract
The rabbit was the initial animal model to investigate the acetylation polymorphism expressed in humans. Use of the rabbit model is compromised by lack of a rapid non-invasive method for determining acetylator phenotype. Slow acetylator phenotype in the rabbit results from deletion of the N-acetyltransferase 2 (NAT2) gene. A relatively quick and non-invasive method for identifying the gene deletion was developed and acetylator phenotypes confirmed by measurement of N- and O- acetyltransferase activities in hepatic cytosols. Rabbit liver cytosols catalyzed the N-acetylation of sulfamethazine (p = 0.0014), benzidine (p = 0.0257), 4-aminobiphenyl (p=0.0012), and the O-acetylation of N-hydroxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (N-OH-PhIP; p = 0.002) at rates significantly higher in rabbits possessing NAT2 gene than rabbits with NAT2 gene deleted. In contrast, hepatic cytosols catalyzed the N-acetylation of p-aminobenzoic acid (an N-acetyltransferase 1 selective substrate) at rates that did not differ significantly (p > 0.05) between rabbits positive and negative for NAT2. The new NAT2 genotyping method facilitates use of the rabbit model to investigate the role of acetylator polymorphism in the metabolism of aromatic and heterocyclic amine drugs and carcinogens.
Introduction
The rabbit was the initial animal model to investigate the acetylation polymorphism expressed in humans (reviewed in Weber and Hein, 1985). Like humans, rabbits express two N-acetyltransferase isozymes, N-acetyltransferase 1 (NAT1) and 2 (NAT2) (Vatsis et al., 1995). Use of the rabbit model is compromised (no publications on use of the rabbit for studies of the acetylation polymorphism have been published for decades) by lack of a published method for determining acetylator genotype. Slow acetylator phenotype in the rabbit results from deletion of the N-acetyltransferase 2 (NAT2) gene (Blum et al., 1989; Sasaki et al., 1991). We developed a rapid NAT2 genotype method in the rabbit and confirmed its accuracy for predicting acetylator phenotype through measurement of hepatic N-and O-acetyltransferase activities.
Materials and methods
Genomic DNA and snap-frozen liver samples from twelve adult New Zealand White rabbits were purchased and shipped from Pel-Freeze Biologicals (Rodgers, AR).
Rabbit NAT2 contains a single open reading frame of 870 nucleotides encoding a 290 amino acid protein (Blum et al., 1990). The rabbit NAT2 and β-globin genes were amplified by polymerase chain reaction (PCR). The primers for the rabbit NAT2 gene were 5’-ccacaatgttaggagggtttgtttat-3’ and 5’-aaggaacaaattgatttgtctaaaaacac-3’ (674-647). The primers for the rabbit β-globin gene were 5’-cccagaggttcttcgagtcctt-3’ (518-539) and 5’-gtgcagcttgtcacagtgcagt-3’. The PCR reaction (20 µL) contained 10 mM Tris-HCL, 50 mM KCL, 3.0 mM MgCl2, 0.2mM of each dNTP, 0.2 µg of each rabbit NAT2 gene primer, 0.008 µg of each rabbit β-globin gene primer, 0.6 U Taq DNA Polymerase (PE Biosystems, Foster City CA) and 10–100 ng of rabbit genomic DNA. Samples were incubated at 94°C for 5 min, followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min. The reaction was concluded with incubation at 72°C for 5 min. PCR products were separated on a 2% agarose gel and visualized under UV light.
Individual cytosols (100,000×g) were prepared as previously described (Fretland et al., 2003) from livers of seven rapid and five slow acetylator rabbits identified by the NAT2 genotype method described above. The rabbit liver cytosols were tested for N-acetyltransferase activities towards p-aminobenzoic acid (PABA) selective for rabbit NAT1 and sulfamethazine (SMZ) selective for NAT2 and two aromatic amine carcinogens: 4-aminobiphenyl (ABP) and benzidine (BZD). The cytosols also were tested for their ability to activate N-hydroxy-2-amino-1-methyl-6-phenylimidazo [4,5-b] pyridine (N-OH-PhIP) by O-acetylation. Substrate concentrations were 30 µM for PABA and SMZ and 100 µM for ABP, BZD and N-OH-PhIP. Acetyl coenzyme A concentration was 1 mM in all assays. Substrates and acetylated products (both mono and di-acetylated products of BZD) were separated and quantitated by high performance liquid chromatography as previously described (Zenser et al., 1996; Fretland et al., 2003; Hein et al., 2006).
Hepatic N-acetyltransferase activities are presented as Mean ± SEM (Figure 2) and individual data scatter plots (Figure 3). Differences in N-acetyltransferase activities between rabbits positive and negative for the NAT2 gene were tested for significance by Student’s T test.
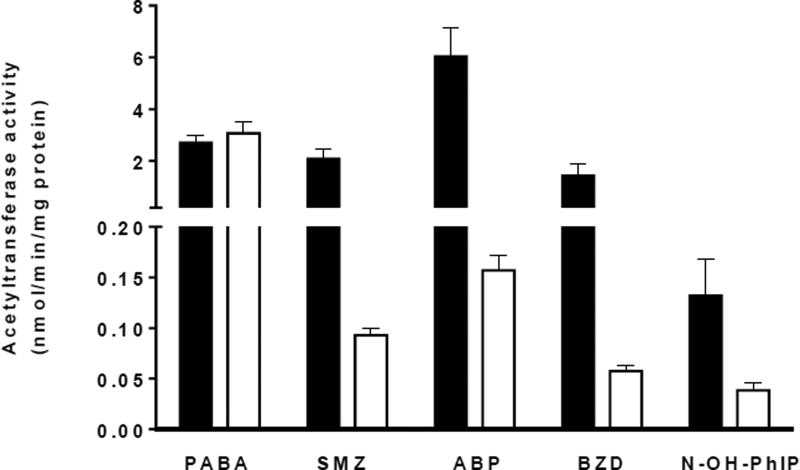
Figure 2.
Acetyltransferase activities in rabbit liver cytosols. Bars represent Mean ± SEM for seven rabbits positive for the NAT2 gene (black) and five rabbits with NAT2 gene deletion (white). SMZ, ABP, and BZD N-acetyltransferase and N-OH-PhIP O-acetyltransferase activities were significantly (p < 0.05) higher in rabbits positive for the NAT2 gene versus rabbits without the NAT2 gene whereas PABA N-acetyltransferase activities did not differ significantly (p>0.05).
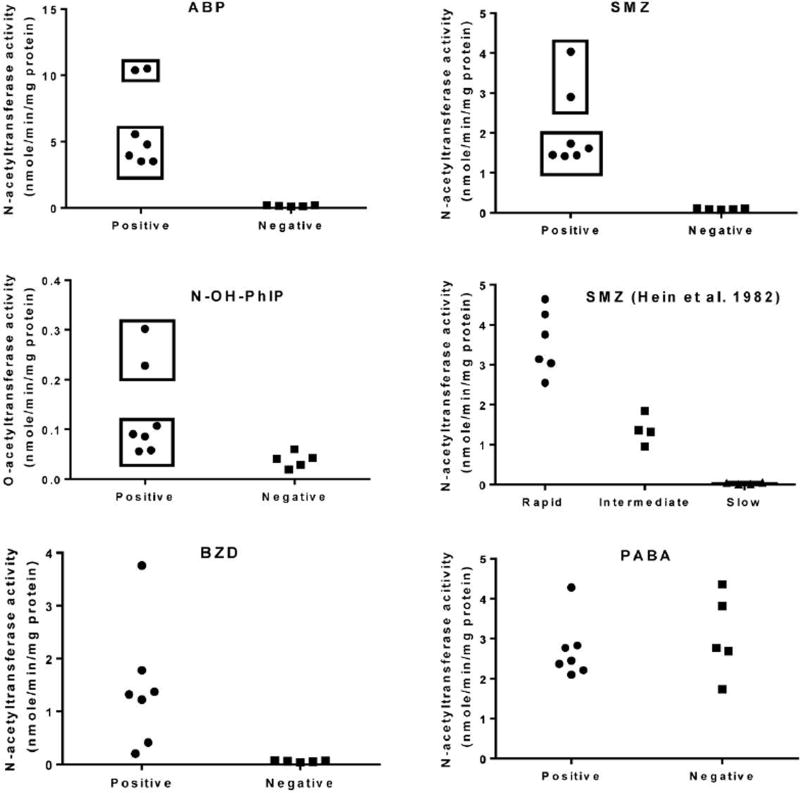
Figure 3.
Scatter plots of ABP, SMZ, BZD and PABA N-acetyltransferase activities in liver cytosols of rabbits positive and negative for presence of the NAT2 gene (the data is presented as Mean ± SEM in Figure 2). Rabbit liver cytosolic SMZ N-acetyltransferase activities from homozygous rapid, heterozygous intermediate, and homozygous slow acetylators redrawn from a previous publication (Hein et al., 1982) also are illustrated for comparison. Based on this comparison, the top rectangles illustrate N-acetyltransferase activities for two rabbits that likely possess two copies of the NAT2 gene whereas the bottom rectangles illustrate N-acetyltransferase activities for five rabbits that likely possess a single copy of the NAT2 gene.
Results and Discussion
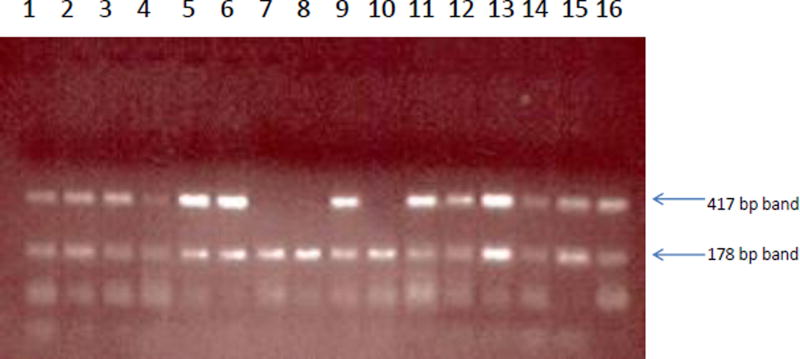
As shown in Figure 1, rabbits possessing the rabbit NAT2 gene (rapid acetylators) had a band of 417 bp whereas rabbits with a deletion of the rabbit NAT2 gene (slow acetylators) did not. All rabbits showed a band of 178 bp for the rabbit β-globin gene which served as the positive control.
Figure 1.
Rabbit NAT2 genotyping method. Samples possessing the rabbit NAT2 gene (Lanes 1–6, 9, 11–16) had a band of 417 bp whereas rabbits with a deletion of the rabbit NAT2 gene (Lanes 7, 8 and 10) did not. All rabbits showed a band of 178 bp for the rabbit β-globin gene which served as the positive control.
SMZ is selective for NAT2 whereas PABA is selective for NAT1 in both humans and rabbits (reviewed in Vatsis et al., 1995). Evidence has been reported that rabbit liver NAT2 catalyzes both N- and O-acetylation (Glowinski et al., 1980). As shown in Figure 2, rabbit liver cytosols catalyzed the N-acetylation of SMZ (p = 0.0014), BZD (p = 0.0257), ABP (p=0.0012), and the O-acetylation of N-OH-PhIP (p = 0.002) at rates significantly higher in rabbits possessing NAT2 gene than rabbits with NAT2 gene deleted. In contrast, hepatic cytosols catalyzed the N-acetylation of PABA (an N-acetyltransferase 1 selective substrate) at rates that did not differ significantly (p > 0.05) between rabbits with or without the NAT2 gene. The magnitude of the difference for N-hydroxy-PhIP O-acetyltransferase activity was considerably lower than for SMZ, ABP, and BZD, suggesting relatively less selectivity for rabbit NAT2 versus NAT1 than SMZ, ABP, and BZD.
The genotyping assay distinguishes rabbits with NAT2 gene deletion, but does not distinguish between rabbits with one or two copies of the NAT2 gene. Previous studies in the rabbit reported trimodal distributions of N-acetyltransferase activity corresponding to homozygous rapid, heterozygous intermediate and homozygous slow acetylators (Hein et al., 1982) which we have redrawn as a scatter plot in Figure 3. We compare this previous scatter plot data with the data obtained in the current investigation also presented as scatter plots (Figure 3). Based on these comparisons, particularly with respect to SMZ, it is likely that 2 of the rabbits positive for the NAT2 gene have two copies (i.e., are homozygous rapid acetylators) whereas 5 of the rabbits positive for the NAT2 gene have a single copy (i.e., are heterozygous intermediate acetylators). This is particularly evident for N-acetylation with SMZ and ABP and the O-acetylation of N-OH-PhIP. It is not as evident with the N-acetylation of BZD, perhaps because this assay measures the acetylation of two amino groups, and it is not at all evident with PABA which is known to be selective for rabbit NAT1 (Hein et al., 1982).
In summary, a relatively quick and non-invasive method for identifying the NAT2 gene deletion in rabbit was developed and acetylator phenotypes confirmed by measurement of N- and O- acetyltransferase activities in hepatic cytosols. The new NAT2 genotyping method facilitates use of the rabbit model to investigate the role of acetylator polymorphism in the metabolism of aromatic and heterocyclic amine drugs and carcinogens.
References
- Blum M, Grant DM, Demierre A, Meyer UA. N-acetylation pharmacogenetics: a gene deletion causes absence of arylamine N-acetyltransferase in liver of slow acetylator rabbits. Proc Natl Acad Sci U S A. 1989;86(23):9554–7. doi: 10.1073/pnas.86.23.9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, Heim M, Meyer UA. Nucleotide sequence of rabbit NAT2 encoding polymorphic liver arylamine N-acetyltransferase (NAT) Nucleic Acids Res. 1990;18(17):5295. doi: 10.1093/nar/18.17.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fretland AJ, Devanaboyina US, Doll MA, Zhao S, Xiao GH, Hein DW. Metabolic activation of 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine in Syrian hamsters congenic at the N-acetyltransferase 2 (NAT2) locus. Toxicol Sci. 2003;74(2):253–9. doi: 10.1093/toxsci/kfg133. [DOI] [PubMed] [Google Scholar]
- Glowinski IB, Weber WW, Fysh JM, Vaught JB, King CM. Evidence that arylhydroxamic acid N,O-acyltransferase and the genetically polymorphic N-acetyltransferase are properties of the same enzyme in rabbit liver. J Biol Chem. 1980;255(16):7883–90. [PubMed] [Google Scholar]
- Hein DW, Doll MA, Nerland DE, Fretland AJ. Tissue distribution of N-acetyltransferase 1 and 2 catalyzing the N-acetylation of 4-aminobiphenyl and O-acetylation of N-hydroxy-4-aminobiphenyl in the congenic rapid and slow acetylator Syrian hamster. Mol Carcinog. 2006;45(4):230–8. doi: 10.1002/mc.20164. [DOI] [PubMed] [Google Scholar]
- Hein DW, Smolen TN, Fox RR, Weber WW. Identification of genetically homozygous rapid and slow acetylators of drugs and environmental carcinogens among established inbred rabbit strains. J Pharmacol Exp Ther. 1982;223(1):40–4. [PubMed] [Google Scholar]
- Sasaki Y, Ohsako S, Deguchi T. Molecular and genetic analyses of arylamine N-acetyltransferase polymorphism of rabbit liver. J Biol Chem. 1991;266(20):13243–50. [PubMed] [Google Scholar]
- Vatsis KP, Weber WW, Bell DA, et al. Nomenclature for N-acetyltransferases. Pharmacogenetics. 1995;5(1):1–17. doi: 10.1097/00008571-199502000-00001. [DOI] [PubMed] [Google Scholar]
- Weber WW, Hein DW. N-acetylation pharmacogenetics. Pharmacol Rev. 1985;37(1):25–79. [PubMed] [Google Scholar]
- Zenser TV, Lakshmi VM, Rustan TD, et al. Human N-acetylation of benzidine: role of NAT1 and NAT2. Cancer Res. 1996;56(17):3941–7. [PubMed] [Google Scholar]