Abstract
Many proteins fold in apparent two-state behavior, as partially folded intermediates only transiently accumulate and easily escape detection. Besides a native form and a mainly unfolded form, we captured a partially unfolded form of an acyl carrier protein from Micromonospora echinospora (meACP) in the folding/unfolding equilibrium using chemical exchange saturation transfer NMR experiments. The C-terminal region of the partially unfolded form is mainly folded and the N-terminal is unfolded. Furthermore, to understand how the folding process of meACP is influenced by solvent environments, we compared the folding dynamics of meACP in D2O, H2O and low concentration of urea. As the environment becomes more denaturing from D2O to H2O and then to urea, the unfolded state becomes increasingly populated, and the folding rate decreases. Adding a small amount of urea, which does not change solvent viscosity, has little effects on the unfolding rates, while changing H2O to D2O reduces the unfolding rates possibly due to the increase of solvent viscosity. The quantified solvent effects on the protein folding Gibbs energy and activation energy suggest that the transition state of folding may have a similar structure to the native state of the protein.
Introduction
Protein molecules exist in multiple conformational states under native conditions. The dominant folded state with the lowest free energy is in dynamic equilibrium with minor states such as unfolded state and partially unfolded intermediate states1–6. Studying the conformational exchanges among different states of a protein can reveal structure-function relationship6–8 or/and help to understand protein folding3,4,9.
Protein folding to biologically functional conformations is important to all living organisms. Misfolding may lead to fatal diseases. Our understanding of protein folding has advanced in the past half-century, yet a general mechanism that applies to most proteins remains missing10. The earliest classic view that proteins fold non-randomly through intermediate states on “predetermined” folding pathways, came from the famous Levinthal paradox that was proposed in 196911. Later studies have well demonstrated the existence of folding intermediates, on-pathway and off-pathway. Different models such as the framework model12, the diffusion-collision13, and the jigsaw puzzle model14 were proposed based on experimental and theoretical studies. However, a consensus has not been reached yet; some models were even contradictory to each other. The “folding funnel energy landscape” view, proposed in the 1990’s, emphasizes the ensemble nature of protein conformations, describing macroscopic “states” as a distribution of various conformations during parallel folding events15. In this “funnel” theory, the so-called off-pathway states are energy traps that are direct and “on-route” for the population involved in these pathways. In more recent years, different hierarchical hypotheses have been revived, such as the “zipping and assembly”16 and “foldon” views3,17–20. The concept of “foldon”, small cooperative folding unit, emerged since the folding of many proteins was detected to be sub-globally cooperative. The “foldon” view embraces the idea of predetermined macroscopic folding pathways and suggests the sequential nature of “foldon” forming processes.
In most of the views, folding intermediates are the keys to understand the folding processes. Although they have been detected in many protein systems, obtaining their specific structural information has been challenging, as high energy intermediate states are often transiently formed and sparsely populated in the native unfolding-folding equilibrium21. The situation is changing now. The advancement of powerful analytical techniques in recent years such as nuclear magnetic resonance (NMR)3,22–24 and mass spectrometry9 has made it possible to characterize the intermediate states at high temporal and spatial resolution.
Shifting protein folding equilibrium by co-solvents such as heavy water (D2O) and urea has been well characterized in the literature. Most of the protein molecules are more stable (Table S1) and more rigid25 and fold more rapidly26 in D2O than in H2O, because of stronger D-bond than H-bond in neutral water27. Urea is a widely used denaturant although its mechanism of denaturing remains not clearly known. Studies suggest that urea denatures proteins by a “direct interaction” mechanism, as urea has preferential binding to all regions of proteins and can form H-bond with protein backbones28,29.
Recently we have found that the acyl carrier protein from Micromonospora echinospora (meACP, PDB code: 2l9f) exists in one major native folded state (N), one minor largely unfolded state (U), and one minor intermediate state (I) under native conditions2,30. The three states are in relatively slow conformational exchange on the sub-second timescale, following the pathway N ↔ U ↔ I or U ↔ N ↔ I. Beyond this, the overall structural features of this state I were mostly uncertain. In this study, we deepened our study of meACP folding to explore structural features of state I using chemical exchange saturation transfer (CEST) NMR. We extracted the backbone 13Ca chemical shifts of meACP in the state I, and demonstrated that this state is a partially unfolded form. In an effort to understand the folding dynamics thoroughly, the folding equilibria were compared under varied conditions: D2O, H2O, and low concentration of urea (0.25 M and 0.5 M). The comparison provides insights into the folding of meACP.
Results and Discussions
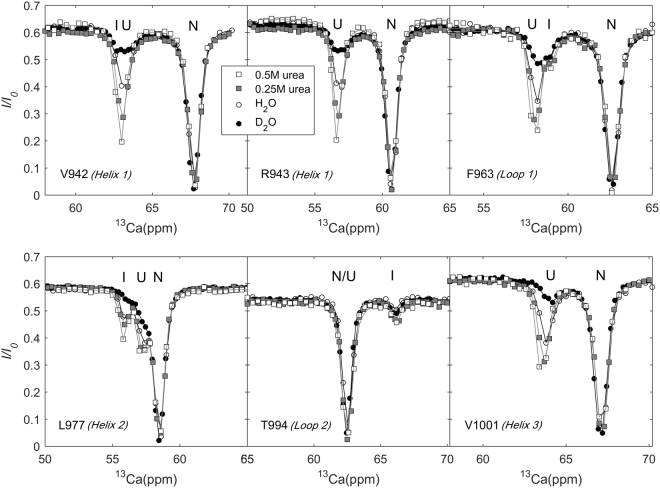
13Ca and 15N CEST experiments on meACP in H2O, D2O, H2O + 0.25 M urea, and H2O + 0.5 M urea were performed. 43 residues displayed two dips in their 13Ca CEST profiles, showing those residues existed in at least two forms. As demonstrated previously on the basis of chemical shifts, the minor form was mainly unfolded (U) with α-helical propensity2,31. Moreover, a third state (I) was clearly seen in their CEST profiles for several residues (Figs. 1 and S1).
Figure 1.
Representative 13Ca CEST profiles obtained with a RF weak field of 15 Hz in H2O, D2O, 0.25 M urea, and 0.5 M urea. The data obtained with a field of 30 Hz are not included for visual clarity. Solid lines are the best fits.
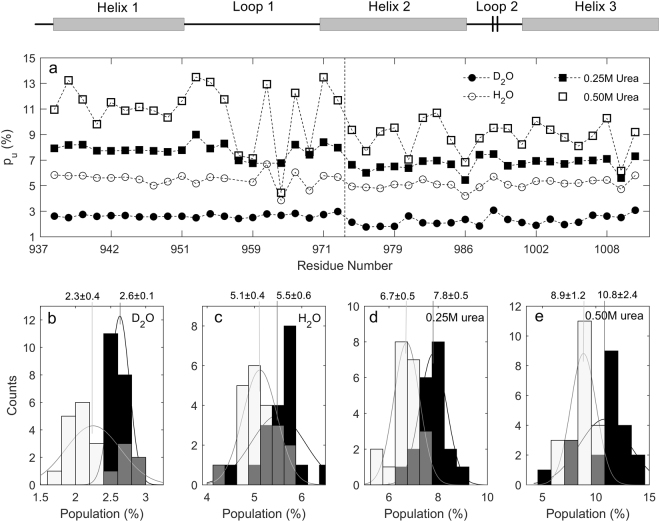
For the residues displaying two-dip CEST profiles, we first performed individual fitting using the two-state model. The population values of state U (p u) extracted from the 13Ca CEST are shown in Fig. 2. The results from the 15N CEST are given in Fig. S4. Interestingly, the average p u values obtained by individual fitting with the two-state model for the residues in the C-terminal half region (C-terminal region of helix 2 and helix 3: from V974 to A1010) were smaller than those for the residues in the N-terminal half region (helix 1, loop 1 and N-terminal region of helix 2: from A938 to I972). The t-tests (p-value < 1.4% for all data sets) confirmed that the differences were significant. The population differences between the two regions showed that the folding of meACP was not globally all or none. Is there a partially folded state where the C-terminal region remains folded while the N-terminal region is unfolded?
Figure 2.
Population of state U p u (a), p u distribution of the two half regions (N-terminal: black bar; C-terminal: white bar; overlapped portion of black and white bars: grey bar) of meACP in D2O (b), H2O (c), 0.25 M urea (d) and 0.5 M urea (e). The averages and standard deviations of pu for the two halves in different solvents are shown on the top of each panel.
When the dip for state I overlaps with that for state U (i.e., ) or overlaps with that for state N (), the CEST profiles of a three-state system look like two-state profiles, but cannot be fitted well to the global two-state model. The bad global fitting of our data by the two-state model indicates the global presence of state I (Fig. S2). In addition, 18 (out of 93) residues displayed three-dip 15N CEST profiles with clear evidence of a third state I2. Moreover, the dip depth for state I in CEST profiles increased with urea concentration (Figs. 1 and S1), and the difference of p u values between the N- and C-terminal regions increased as well (Fig. 2). Taken together, we suggest that the difference of p u between the two regions shown in Fig. 2 comes from the global presence of state I.
For a system with three conformational states, there are four possible three-state exchange models (M2~M5 in Fig. 3). As discussed in our previous paper2, the exchange rates between N and U and between N and I were around 250 s−1 and 100 s−1, respectively, and model M2 was excluded based on the 15N data. In the on-pathway model (M2 in Fig. 3), the exchange between N and I must be faster than the exchange between N and U, which is contradictory to the result derived from the 15N data. It is noteworthy that models M3 and M4 are two special cases of M5, where the exchange rate between I and N or the rate between I and U is zero.
Figure 3.
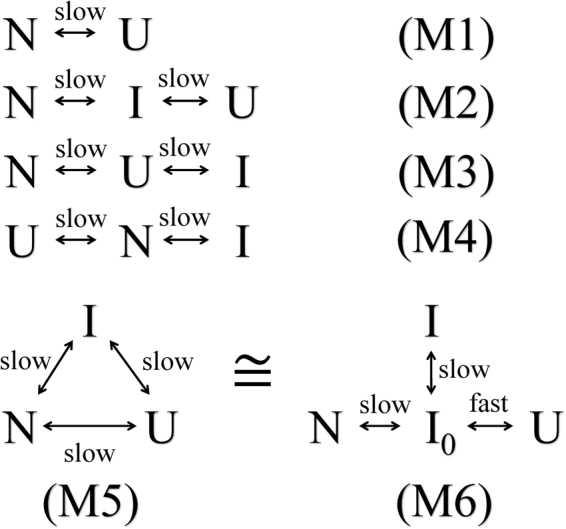
Possible exchange models examined for meACP.
For each data set, global fittings based on the two-state model and three-state models were conducted to compare the fitting goodness, and to extract the global exchange rates and populations of different states (shared by all residues) as well as the per-residue chemical shifts of minor states (ΩU only in two-state model; ΩU and ΩI in three-state models), transverse relaxation rates (R2N) and longitudinal relaxation rates (R1N) of the major state. The reduced x 2 values for global fittings by three possible three-state models M3~M5 were significantly smaller than those for the two-state model M1 and the on-pathway three-state model M2 (see Table S2). The F-tests showed that the global three-state models M3~M5 described the CEST data significantly better than models M1 and M2 (p-value < 10−4 for all data sets). Indeed, the two-state model failed obviously for a few residues that displayed three dips. Considering the possibility that global minima might not be reached in the global fittings by the three-state models (due to large search space) and the fact that the fitting residuals by M3~M5 were similar, we think that the models M3~M5 matched equally well with the data (Table S2). So we could not determine which model is true for meACP. Fortunately, the 13Ca chemical shifts of state I (ΩI) obtained with these three three-state models were almost identical.
As the ΩI values were extracted by minimizing χ2, it is likely that ΩI could be positioned incorrectly at ΩU in a few cases when ΩI and ΩU were very similar and χ2 values were not sensitive to ΩI positions. Nevertheless, ΩI could be determined at high confidence level from the χ2 distribution for most residues.
Structural features of the two minor states
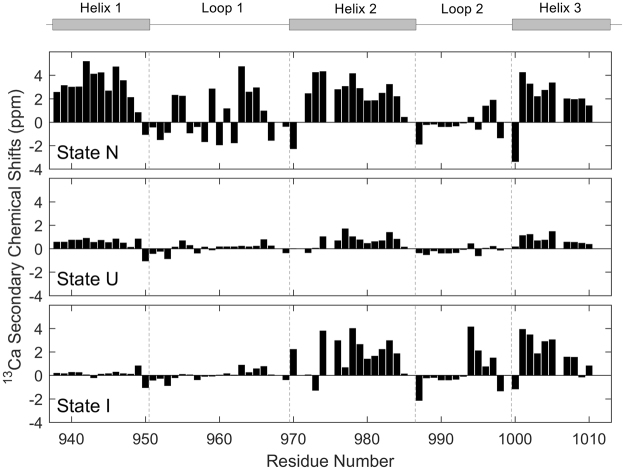
The secondary chemical shifts (CSs) of each residue in three states were calculated by subtracting the random coil chemical shift (CS) values, which were experimentally measured in the fully denatured sample (meACP in 4 M urea), from the CS values of the corresponding residues which were obtained in the CEST data. The analysis of 13Ca secondary CSs is a simple method to identify the secondary structures of a protein in different states32 (Fig. 4).
Figure 4.
13Ca secondary chemical shifts of state N, U and I in D2O. 13Ca chemical shifts experimentally measured in 4 M urea (fully denatured) were used as the reference.
For state U, the 13Ca secondary CSs in three helical regions had an average value of around +1 ppm, as compared with state N whose 13Ca secondary CSs in the helical region were in the range of +2~+3 ppm (Fig. 4), indicating the presence of residual helical structures. The residual helical pattern in state U was the same as that in the native state, implying that the three native helices were not fully unfolded in state U. The 13Ca secondary CS amplitudes for the helical regions in state U were about one-third of those in state N, suggesting that the three helices in state U are populated for about 30% of the time.
For state I, the 13Ca secondary CSs of its N-terminal region (helix 1, loop 1, and N-terminal region of helix 2) were close to zero, indicating this region exists in a nearly fully unfolded form. On the other hand, the 13Ca secondary CSs of the C-terminal region (helix 3 and C-terminal region of helix 2) in state I were mostly similar to those in state N, showing that the C-terminal region of state I is native-like (Fig. 4) (or the helices 2 and 3 remain mainly folded in state I).
Obviously, the state I is overall a partially unfolded form (PUF). This is consistent with the results shown in Fig. 2. Based on the two-state model, a residue can stay as either a folded or an unfolded form. As the N-terminal region has a higher unfolded population than the C-terminal region (Fig. 2), some protein molecules must exist as a PUF. Based on 15N CEST data alone, we previously suggested that state I adopted a locally altered conformation in which the N-terminal region of helix 2 differs from both helical and random coil structures while other helices are native-like2. Based on 13Ca and 15N data, it is now clear that the state I resembles state N in the C-terminal half, while it resembles state U in the N-terminal half (Fig. 4). It is noteworthy that state I still differs significantly from state N in the N-terminal region of helix 2.
The partially unfolded form (PUF or state I)
In one scenario, the folding follows the triangle model (M5 in Fig. 3). It might suggest parallel folding pathways. The terms p I * k IN and p u * k UN can represent the contributions to folding from apparent folding pathways U-I-N and U-N, respectively. was estimated to be smaller than ~0.06 for all data sets (Table S3), showing that the pathway U-I-N contributes significantly less than the U-N pathway in the protein folding. Although the triangle model fits the experimental data, it is not necessary for the folding to must be parallel, because a four-state model with state I as an off-pathway product (M6 in Fig. 3) still fits the data. Therefore, the state I might be an “on-pathway” folding intermediate state and it is also possible to be an “off-pathway” intermediate state.
The triangle model can be approximated as the single-pathway model N-U-I (M3) when k IN + k NI is much smaller than k IU + k UI. It can also be approximated as the model U-N-I (M4) when k IN + k NI is much larger than k IU + k UI. For some of our data sets k IN + k NI ≫ k IU + k UI, and for other data sets k IN + k NI ≪ k IU + k UI (Table S3). This is caused by the fact that each set of data could be fitted equally well by M3 and M4 (Table S2). Because the folding can follow the two possible single-pathway models, state I might be an “off-pathway” state of the folding from state U to N. We previously suggested that there might be a dynamic equilibrium between monomeric and oligomeric forms, and the oligomers might result from this state I2. The N-terminal region of helix 2 (around S970-L977) in state I is very non-native (Fig. 4), which might be prone to aggregate. The misfolding of this region might cause the failure of the further folding of state I.
If state I0 is on the folding path from U to I0 to N, but local misfolding happens for a fraction of this state and blocks its folding to state N due to “the occurrence of an on-pathway optional error”33, the misfolded fraction (state I) appears as an off-pathway product (“dead-end”) (M6 in Fig. 3). Therefore, the “off-pathway” state I may represent the misfolded population, which is only a fraction of the constructive state I0 with optional local misfolding errors. This putative “on-pathway” intermediate state I0 would be similar to the observed state I in overall structure, but it would interconvert with state U so fast that it could not be detected by CEST and relaxation dispersion.
Comparison of folding dynamics in D2O, H2O, and urea
The effects of co-solvent D2O and urea on shifting the folding equilibria were examined. The 13Ca CEST profiles recorded in H2O, D2O and urea varied mainly in the depths of the minor dips (Fig. 1). The dips for both states U and I were larger in urea and smaller in D2O, compared with those in H2O (see Fig. 1 and Fig. S1).
Table 1 shows the extracted folding parameters in D2O, H2O, 0.25 M urea and 0.50 M urea by following M3 (N-U-I). Models M4 and M5 gave similar p u, k UN, k NU, ΩU, and ΩI values as M3 did (see Tables S3 and S4). As buffer condition became more denaturing (from D2O to H2O, to higher concentration of urea), the population of unfolded state (p u ) increased, reflecting the decrease of Gibbs energy of unfolding (ΔGN–U). We do expect unfolded states (U) to be more populated in urea, less in D2O, as D2O and urea have been widely known for their stabilizing and destabilizing effects on proteins, respectively. meACP was ~0.5 kcal/mol more stable in D2O (Table 1). This is consistent with literature studies (summarized in Table S1). Previous data suggest that the larger the proteins are, the more prominent the stabilizing effect is (Fig. S3). The stability enhanced by D2O agrees well with the size of meACP.
Table 1.
Folding and unfolding based on three-state model N-U-I.
| Solvents | Expt. | pu (%) | pI (%) | ΔGNU (kcal/mol) | ΔGNI (kcal/mol) | kNU (s−1) | kUN (s−1) | kUI + kIU (s−1) |
|---|---|---|---|---|---|---|---|---|
| D2O | 13Ca | 2.0 ± 0.01 | ~2.0* | 2.3 | ~2.3* | 10.4 ± 2.3 | 495.2 ± 7.9 | ~73* |
| H2O | 13Ca | 4.6 ± 0.02 | ~1.9 | 1.8 | ~2.3 | 16.9 ± 2.6 | 356.6 ± 10.4 | ~141 |
| H2O | 15N | 4.8 ± 0.01 | ~0.8 | 1.8 | ~2.8 | 15.0 ± 0.3 | 297.6 ± 4.9 | ~85 |
| 0.25 M Urea + H2O | 13Ca | 6.5 ± 0.04 | ~2.1 | 1.6 | ~2.2 | 14.9 ± 0.8 | 220.0 ± 5.5 | ~93 |
| 0.25 M Urea + H2O | 15N | 6.9 ± 0.05 | ~6.9 | 1.5 | ~1.5 | 15.7 ± 0.3 | 199.6 ± 2.7 | ~34 |
| 0.50 M Urea + H2O | 13Ca | 7.9 ± 0.04 | ~8.5 | 1.4 | ~1.4 | 15.6 ± 0.6 | 176.5 ± 4.1 | ~31 |
*, “~” is used to indicate that the results were estimated as p I and (k UI + k IU) are correlated in the fitting.
The exchange between states N and U can be viewed as the major folding/unfolding process, therefore the conversion rates from U to N (k UN) and from N to U (k NU) were denoted as the folding (k f) and unfolding (k u) rates, respectively. According to the results in Table 1, Tables S3 and S4, k f (k UN) decreased as solvent became more denaturing; k u (k NU) increased when solvent was changed from D2O to H2O, and there were no significant effects on k u by a low concentration of urea.
There were large uncertainties in the population (p I) and exchange rates of state I (k X–I and k I–X, which are the exchange rates between I and N and between I and U) because the fitting x 2 values changed little when p I, k X–I, and k I–X were within certain ranges. Therefore, we only estimated p I and the sum of k X–I and k I–X. Despite the large uncertainty in the estimated p I, we observed in the raw profiles that the dips for state I were larger in urea, and smaller in D2O than in H2O (Figs. 1 and S1). The variations of state I dips in the raw profiles indicate that the population of state I increases with the destabilization strength of the solvent (in current range of low concentrations of urea).
Effects of urea on meACP folding and unfolding
High concentration of urea is denaturing, while the effects of low concentration of urea on protein stability seem to vary for different proteins and solvent environments. It was shown that low concentration urea stabilizes protein ferrocytochrome c34. Here we found that meACP was destabilized, even at the concentration of urea as low as 0.25 M. The populations of both state U and state I increased with urea concentration.
According to Eq. 4 (see Materials and Methods section), the effect of urea on k f mainly originates from the change of energy barrier (activation energy ), because the changes of viscosity and internal friction at low concentration of urea buffer (0.25 M) are negligible ()35,36. Therefore, the change of activation energy is given by,
| 1 |
Using the results in Table 1, was estimated as ~0.29 kcal/mol and ~0.42 kcal/mol for meACP at 0.25 M and 0.5 M urea, respectively (Fig. 5). Similarly, (the change of the Gibbs energy of folding, ) was calculated from extracted p u values with Eq. 3. was ~0.25 kcal/mol and ~0.40 kcal/mol for meACP in 0.25 M and 0.5 M urea (relative to H2O), respectively (Fig. 5). Accordingly,
| 2 |
Figure 5.
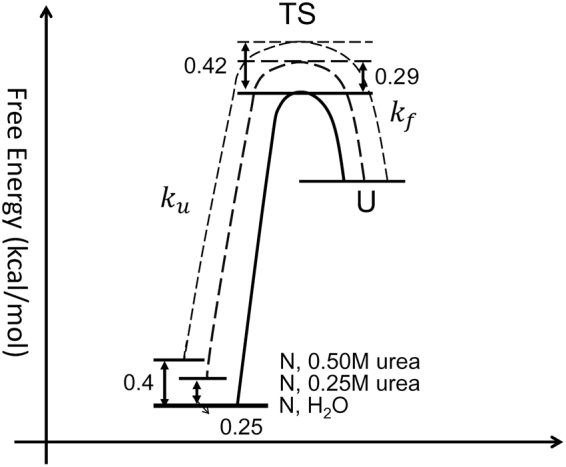
Schematic plot of relative free energy profiles of meACP folding in varied solvents. The free energies in states TS and N are relative to that in state U. (Energy values were calculated based on Table 1).
The ϕ f factor, or so-called Tanford’s β value, is viewed as an indicator of the average degree of changes in accessible surface areas (ΔASA) of the transition state (TS) relative to that of the unfolded state during the protein folding37. ϕ f, generally ranging from 0 to 1, represents the ratio of TS stability change to native state (N) stability change. A ϕ f value of 1, meaning that the relative stability change of TS equals to that of N, suggests that the overall ΔASA of TS and that of N are similar during unfolding. Thus, indicates that the TS may have a native-like structure. According to the results obtained in D2O, we found that ϕ f > 1/2, further supporting that the TS is native-like (see discussion in the supplementary information).
Overall, the results here suggest that the TS could be placed close to state N on the path N ↔ U in the possible models (M3~M6) shown in Fig. 3. In M3, the pathway would be N ↔ TS ↔ U ↔ I when considering the TS; in M4 it would be U ↔ TS ↔ N ↔ I; in M5 it would be N ( ↔ I) ↔ TS ↔ U ( ↔ I); in M6 it would be U ↔ I0 ( ↔ I) ↔ TS ↔ N, according to their structural features (or overall folded percentages). The TS is suggested to be a folded transition state that is close to the native state, sharing a similar structure with the native state.
Conclusion
We found that there exists a partially unfolded form of meACP, where the N-terminal half is unfolded while the C-end half remains mainly folded. The existence of PUF was confirmed by 15N and 13Ca CEST results in different solvents. The detected PUF could be either an “on-pathway” intermediate state in the triangle model or an “off-pathway” state in the other three-state models and four-state model. This PUF might represent a fraction of the constructive folding intermediate, and occurred to be misfolded. The structure features of this PUF might suggest the sequential folding order in which the C-terminal region folds before N-terminal region does. Furthermore, the H/D isotope effect and urea effect on meACP folding dynamics were investigated by comparing CEST results in different solvent conditions. Consistent with literature studies on H/D isotope effects, the native state of meACP was more stable in D2O, folding was accelerated, and unfolding was decelerated compared to that in H2O. Low concentration of urea destabilized the native state of the protein and seemed to stabilize state PUF. It also slowed down the folding rates while had no effects on the unfolding rates, suggesting the structural similarity between the transition state and native state of meACP.
Materials and Methods
Sample preparation and NMR spectroscopy
Different acyl carrier protein samples (Table 2) were prepared by following previous protocols38. All NMR samples contained 0.6~1 mM protein, 50 mM NaCl, 5 mM EDTA, and 50 mM phosphate at pH 6.9. 13Ca CEST and 15N CEST experiments were performed at 25 °C on a Bruker 800 MHz NMR spectrometer equipped with a cryoprobe. The experimental parameters used were the same as those described previously2,31.
Table 2.
CEST experiments.
| No. | Sample labeling | Solvent Condition | Experiment |
|---|---|---|---|
| 1 | 13C | 95% H2O, 5% D2O | 13Ca CEST |
| 2 | 13C | 100% D2O | 13Ca CEST |
| 3 | 13C | 95% H2O, 5% D2O + 0.25 M urea | 13Ca CEST |
| 4 | 13C | 95% H2O, 5% D2O + 0.50 M urea | 13Ca CEST |
| 5 | 15N | 95% H2O, 5% D2O | 15N CEST |
| 6 | 15N | 95% H2O, 5% D2O + 0.25 M urea | 15N CEST |
CEST data analysis
Individual and global fitting using the two-state model
The residues displaying two dips separated by more than 1 ppm were chosen to extract kinetics parameters. Their CEST profiles were first fitted individually using the two-state model (M1 in Fig. 3) to obtain parameters for each residue, i.e., folding and unfolding rates (k f, k u), population of the minor state (p u), chemical shift in the minor state (ΩU), longitudinal relaxation rates of major (R1N) and minor states (R1U), transverse relaxation rates of the major (R2N) and minor states (R2U). In the fitting, the J coupling effect was taken into account as described previously39. We assumed that 1JCOCα = 55 Hz and 1JCαCβ = 35 Hz for all residues in 13C labelled protein samples; and set R1U = R1N for each residue. When R2U was used as an independent fitting parameter, the extracted R2U for most residues deviated significantly from the expected values for our system due to the presence of a third state that overlaps with state N or U as well as deviations of 1JCOCα and 1JCαCβ from the assumed values. To simplify the fitting, we assumed all the residues have the same R2U values in the unfolded state and set R2U = 6.5 and 8.1 s−1 for 13Ca in H2O and D2O respectively and 3.0 s−1 for 15N in H2O at 25 °C. These values were estimated based on the 15N R2 values of intrinsically disordered protein α-synuclein (~3.5 s−1), which were measured at 15 °C on an 800 MHz spectrometer40. The same assumption was made for three-state fittings. In global fitting, all residues shared a common exchange rate (k ex) and a common population of the minor state (p u), but they each had unique R 1N, R 2N, and ΩU values.
Individual fitting and global fitting using three-state models
The procedures for fitting the CEST data of individual residues to three-state models (M2 ~ M5) were the same as that described above. The residues displaying two and three dips were also fitted globally to the three-state models. In the fitting, we set R 2I = R 2N and R 1I = R 1U = R 1N for each residue, and assumed 1JCOCα = 55 Hz and 1JCαCβ = 35 Hz for all residues. For residues displaying three well separated dips, ΩU and ΩI values were already certainly known. For residues displaying two-dip profiles, ΩU values were quite certain and ΩI should be close to either ΩN or ΩU. Optimization was done by extensive grid-search of ΩI. To extract the ΩI value as accurate as possible, we used the following procedure:
The ΩI of each residue was first estimated by individual fitting via grid search of ΩI;
ΩI values obtained in step a were used as the input values in the global fitting to obtain global exchange rates between states N and U (k1) and between I and U or I and N (k2), populations of states N (p N), and U (p u);
ΩI values were re-calculated by individual fitting with fixed global k1, k2, p N, and p u values (obtained in step b);
Repeat steps b and c, until χ2 of the global fitting decreased to a stable value.
Error estimation
For each CEST profile, the uncertainty (δ) in intensity ratio (I/ I 0) was estimated by calculating the standard deviation of data points in the ‘non-saturation area’. The following Monte Carlo simulations were used to estimate fitting errors of extracted CEST parameters for each residue:
Generate 120 sets of profiles using extracted parameters, add random noise with a standard deviation of δ and mean of 0;
The 120 set of profiles were fitted to extract 120 sets of fitting parameters. The standard deviation of each parameter was considered as the fitting error.
To determine global fitting errors, 80% of the residues used in the global fitting were randomly taken to extract global parameters and repeat 120 times to obtain standard deviations.
Energy calculations
According to the population of each state, the Gibbs energy change (ΔG) of process A to B was calculated by
| 3 |
where p A and p B are the populations of states A and B, respectively. The subscript AB is short for A → B.
The folding rate (k f) is related to the activation energy () by41,42
| 4 |
where C is the frequency factor for the folding process, σ refers to the “internal friction”, and η is the viscosity of solvent. σ reflects the contribution of the energy landscape ruggedness to the reaction rate and is dominated by the structure of a protein when denaturant concentrations are low36.
Data Availability
The datasets used in the current study are available in the supplementary information file, and the Matlab scripts for data fitting are available from the corresponding author upon request.
Electronic supplementary material
Acknowledgements
This work was supported by a Grant from Singapore Ministry of Education Academic Research Fund Tier 3 (MOE2012-T3-1-008).
Author Contributions
D.Y. designed research; Y.Z. performed research; Y.Z. and D.Y. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16449-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vallurupalli P, Bouvignies G, Kay LE. Studying “invisible” excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc. 2012;134:8148–8161. doi: 10.1021/ja3001419. [DOI] [PubMed] [Google Scholar]
- 2.Lim J, Xiao T, Fan J, Yang D. An Off‐Pathway Folding Intermediate of an Acyl Carrier Protein Domain Coexists with the Folded and Unfolded States under Native Conditions. Angew. Chem. 2014;53:2358–2361. doi: 10.1002/anie.201308512. [DOI] [PubMed] [Google Scholar]
- 3.Maity H, Maity M, Krishna MM, Mayne L, Englander SW. Protein folding: the stepwise assembly of foldon units. Proc. Natl. Acad. Sci. USA. 2005;102:4741–4746. doi: 10.1073/pnas.0501043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollen YJ, Kamphuis MB, van Mierlo CP. The folding energy landscape of apoflavodoxin is rugged: hydrogen exchange reveals nonproductive misfolded intermediates. Proc. Natl. Acad. Sci. USA. 2006;103:4095–4100. doi: 10.1073/pnas.0509133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu, B. & Yang, D. Coexistence of multiple minor states of fatty acid binding protein and their functional relevance. Sci Rep6 (2016). [DOI] [PMC free article] [PubMed]
- 6.Xiao T, Fan JS, Zhou H, Lin Q, Yang D. Local Unfolding of Fatty Acid Binding Protein to Allow Ligand Entry for Binding. Angew Chem Int Ed Engl. 2016;55:6869–6872. doi: 10.1002/anie.201601326. [DOI] [PubMed] [Google Scholar]
- 7.Manglik A, Kobilka B. The role of protein dynamics in GPCR function: Insights from the β 2 AR and rhodopsin. Curr. Opin. Cell Biol. 2014;27:136–143. doi: 10.1016/j.ceb.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye, L., Van Eps, N., Zimmer, M., Ernst, O. P. & Prosser, R. S. Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature (2016). [DOI] [PubMed]
- 9.Hu W, et al. Stepwise protein folding at near amino acid resolution by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2013;110:7684–7689. doi: 10.1073/pnas.1305887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dill KA, MacCallum JL. The protein-folding problem, 50 years on. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 11.Levinthal C. Are there pathways for protein folding. J. Chim. Phys. 1968;65:44–45. doi: 10.1051/jcp/1968650044. [DOI] [Google Scholar]
- 12.Udgaonkar JB, Baldwin RL. NMR evidence for an early framework intermediate on the folding pathway of ribonuclease A. Nature. 1988;335:694–699. doi: 10.1038/335694a0. [DOI] [PubMed] [Google Scholar]
- 13.Karplus M, Weaver DL. Protein-folding dynamics. Nature. 1976;260:404–406. doi: 10.1038/260404a0. [DOI] [PubMed] [Google Scholar]
- 14.Harrison SC, Durbin R. Is there a single pathway for the folding of a polypeptide chain? Proc. Natl. Acad. Sci. USA. 1985;82:4028–4030. doi: 10.1073/pnas.82.12.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat. Struct. Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 16.Ozkan SB, Wu GA, Chodera JD, Dill KA. Protein folding by zipping and assembly. Proc. Natl. Acad. Sci. USA. 2007;104:11987–11992. doi: 10.1073/pnas.0703700104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamberlain AK, Handel TM, Marqusee S. Detection of rare partially folded molecules in equilibrium with the native conformation of RNaseH. Nat. Struct. Mol. Biol. 1996;3:782–787. doi: 10.1038/nsb0996-782. [DOI] [PubMed] [Google Scholar]
- 18.Vu N-D, Feng H, Bai Y. The folding pathway of barnase: the rate-limiting transition state and a hidden intermediate under native conditions. Biochemistry. 2004;43:3346–3356. doi: 10.1021/bi0362267. [DOI] [PubMed] [Google Scholar]
- 19.Cellitti J, Bernstein R, Marqusee S. Exploring subdomain cooperativity in T4 lysozyme II: Uncovering the C‐terminal subdomain as a hidden intermediate in the kinetic folding pathway. Protein Sci. 2007;16:852–862. doi: 10.1110/ps.062632807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzkee NC, Torchia DA, Bax A. Measuring rapid hydrogen exchange in the homodimeric 36 kDa HIV‐1 integrase catalytic core domain. Protein Sci. 2011;20:500–512. doi: 10.1002/pro.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sánchez IE, Kiefhaber T. Evidence for sequential barriers and obligatory intermediates in apparent two-state protein folding. J. Mol. Biol. 2003;325:367–376. doi: 10.1016/S0022-2836(02)01230-5. [DOI] [PubMed] [Google Scholar]
- 22.Korzhnev DM, Kay LE. Probing invisible, low-populated states of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding. Acc. Chem. Res. 2008;41:442–451. doi: 10.1021/ar700189y. [DOI] [PubMed] [Google Scholar]
- 23.Bouvignies G, Kay LE. Measurement of proton chemical shifts in invisible states of slowly exchanging protein systems by chemical exchange saturation transfer. J. Phys. Chem. B. 2012;116:14311–14317. doi: 10.1021/jp311109u. [DOI] [PubMed] [Google Scholar]
- 24.Hansen AL, Bouvignies G, Kay LE. Probing slowly exchanging protein systems via 13Cα-CEST: monitoring folding of the Im7 protein. J. Biomol. NMR. 2013;55:279–289. doi: 10.1007/s10858-013-9711-4. [DOI] [PubMed] [Google Scholar]
- 25.Cioni P, Strambini GB. Effect of Heavy Water on Protein Flexibility. Biophys. J. 2002;82:3246–3253. doi: 10.1016/S0006-3495(02)75666-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker MJ, Clarke AR. Amide backbone and water-related H/D isotope effects on the dynamics of a protein folding reaction. Biochemistry. 1997;36:5786–5794. doi: 10.1021/bi9629283. [DOI] [PubMed] [Google Scholar]
- 27.Scheiner S, Cuma M. Relative stability of hydrogen and deuterium bonds. J. Am. Chem. Soc. 1996;118:1511–1521. doi: 10.1021/ja9530376. [DOI] [Google Scholar]
- 28.Lim WK, Rösgen J, Englander SW. Urea, but not guanidinium, destabilizes proteins by forming hydrogen bonds to the peptide group. Proc. Natl. Acad. Sci. USA. 2009;106:2595–2600. doi: 10.1073/pnas.0812588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua L, Zhou R, Thirumalai D, Berne B. Urea denaturation by stronger dispersion interactions with proteins than water implies a 2-stage unfolding. Proc. Natl. Acad. Sci. USA. 2008;105:16928–16933. doi: 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim J, et al. Rigidifying acyl carrier protein domain in iterative type I PKS CalE8 does not affect its function. Biophys. J. 2012;103:1037–1044. doi: 10.1016/j.bpj.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Yang D. 13Cα CEST experiment on uniformly 13C-labeled proteins. J. Biomol. NMR. 2015;61:89–94. doi: 10.1007/s10858-014-9888-1. [DOI] [PubMed] [Google Scholar]
- 32.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 33.Englander SW, Mayne L. The nature of protein folding pathways. Proc. Natl. Acad. Sci. USA. 2014;111:15873–15880. doi: 10.1073/pnas.1411798111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhuyan AK. Protein stabilization by urea and guanidine hydrochloride. Biochemistry. 2002;41:13386–13394. doi: 10.1021/bi020371n. [DOI] [PubMed] [Google Scholar]
- 35.Kawahara K, Tanford C. Viscosity and density of aqueous solutions of urea and guanidine hydrochloride. J. Biol. Chem. 1966;241:3228–3232. [PubMed] [Google Scholar]
- 36.Volk M, Milanesi L, Waltho JP, Hunter CA, Beddard GS. The roughness of the protein energy landscape results in anomalous diffusion of the polypeptide backbone. PCCP. 2015;17:762–782. doi: 10.1039/C4CP03058C. [DOI] [PubMed] [Google Scholar]
- 37.Ramos CH, Weisbuch S, Jamin M. Diffusive motions control the folding and unfolding kinetics of the apomyoglobin pH 4 molten globule intermediate. Biochemistry. 2007;46:4379–4389. doi: 10.1021/bi602574x. [DOI] [PubMed] [Google Scholar]
- 38.Lim J, et al. Solution Structures of the Acyl Carrier Protein Domain from the Highly Reducing Type I Iterative Polyketide Synthase CalE8. Plos One. 2011;6:e20549. doi: 10.1371/journal.pone.0020549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y, Yang D. Effects of J couplings and unobservable minor states on kinetics parameters extracted from CEST data. J Magn Reson. 2014;249C:118–125. doi: 10.1016/j.jmr.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Kim S, Wu KP, Baum J. Fast hydrogen exchange affects ¹υN relaxation measurements in intrinsically disordered proteins. J. Biomol. NMR. 2013;55:249–256. doi: 10.1007/s10858-013-9706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pabit SA, Roder H, Hagen SJ. Internal friction controls the speed of protein folding from a compact configuration. Biochemistry. 2004;43:12532–12538. doi: 10.1021/bi048822m. [DOI] [PubMed] [Google Scholar]
- 42.Ansari A, Jones CM, Henry ER, Hofrichter J, Eaton WA. The role of solvent viscosity in the dynamics of protein conformational changes. Science. 1992;256:1796–1798. doi: 10.1126/science.1615323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in the current study are available in the supplementary information file, and the Matlab scripts for data fitting are available from the corresponding author upon request.