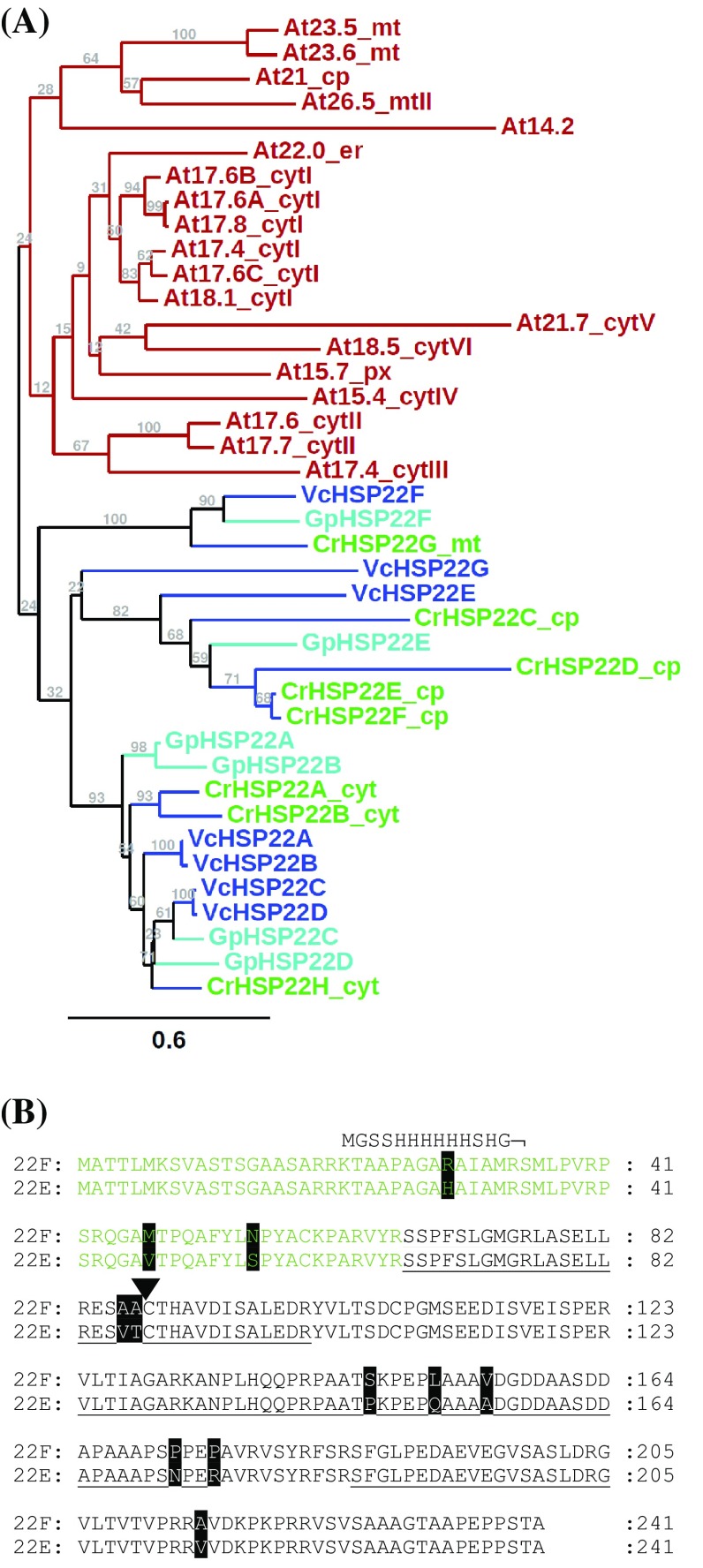
Fig. 1.
Phylogenetic tree of sHsps and comparison of the Chlamydomonas HSP22E and HSP22F amino acid sequences. a Phylogram based on an amino acid sequence alignment of the α-crystalline domains from Arabidopsis thaliana (At) and Volvocales members Chlamydomonas reinhardtii (Cr), Gonium pectorale (Gp) and Volvox carteri (Vc). Protein names are appended by their predicted intracellular localization (cyt cytosol; cp chloroplast; mt mitochondria; er endoplasmic reticulum; px peroxisome) and phylogenetic subfamily (roman numbers) as assigned by Waters et al. (2008) and Schroda and Vallon (2009). Support for the branches is given in bootstrap values based on 1000 NJ bootstrap replicates. b Alignment of HSP22F and HSP22E protein sequences. Underlined sequences indicate peptides identified by LC-MS/MS analysis of the immunoprecipitated proteins (Supplementary Table S3). Sequences shown in green represent the putative chloroplast transit peptide, sequences in black the putative mature protein. The triangle indicates the cleavage site predicted by ChloroP (Emanuelsson et al. 1999). Differences in both sequences are shaded in black. The sequence on top of that of HSP22F shows where the hexahistidine tag is fused to HSP22F in the recombinant protein