Figure 3.
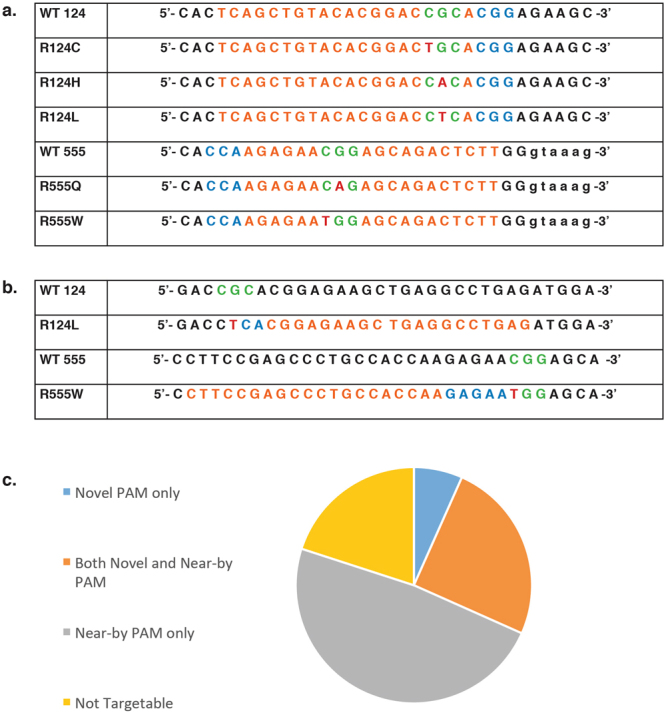
Analysis of TGFBI corneal dystrophy mutations in a CRISPR system Codons 124 or 555 shown in green, mutated base shown in red, nearest PAM to be utilised shown in blue and consequential guide sequence shown in orange. (a) Mutation analysis revealed that none of the prevalent TGFBI mutations generated a novel S. pyogenes PAM, however a naturally occurring PAM exists for all five mutations. For mutations in codon 124 the nearest downstream PAM places the mutated base at either position 3 or 4 of the guide sequence. For mutations in codon 555 the nearest downstream PAM places the mutated base at either position 7 or 8 of the guide sequence. (b) Mutational analysis revealed that R124L generates a novel PAM with a mutant AsCpf1 that recognises a 5′-VYCV-3′ PAM. R124L generates a 5′-CTCA-3′ PAM. Further analysis revealed that R555W generates a novel PAM with S.aureus which is capable of recognising a 5′-NNGRRT-3′ PAM. R555W generates a 5′-GAGAAT-3′ PAM. (c) Venn diagram to illustrate the total number of TGFBI mutations that (i) generate a novel S. pyogenes PAM, (ii) have a near-by S. pyogenes PAM i.e. within the first 8 bp of the guide sequence, (iii) have both a novel and near-by S. pyogenes PAM or (iv) are not targetable by either approach.
