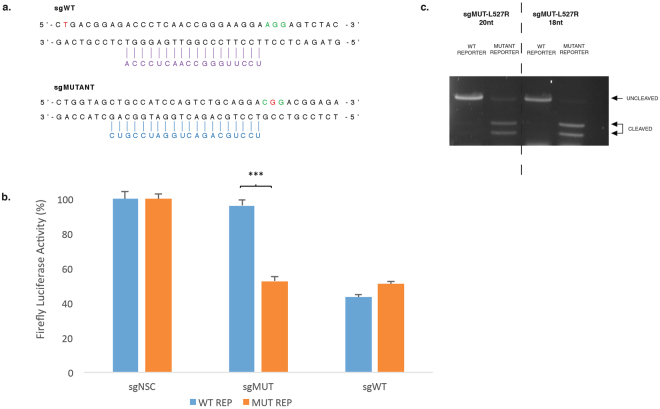
Figure 4.
Allele-specific cleavage of L527R TGFBI mutation utilising a PAM-specific approach. (a) The L527R mutation (c.1580 T > G) is indicated in red and PAM utilised is shown in green. A 20 nt sgRNA targeted to a naturally occurring PAM was designed as a positive control (sgWT, purple –top of figure). A 20 nt sgRNA utilising the novel PAM, containing the L527R mutation, was designed (sgMUTANT, blue – bottom of figure). (b) Both sgWT and sgMUTANT were targeted to a luciferase reporter plasmid containing either a wild-type or mutant TGFBI sequence to determine potency and allele specificity. (c) An in vitro digestion with Cas9 protein complexed with a sgRNA utilising the novel L527R PAM was carried out to confirm the specificity observed. Mutant guides of both 20 and 18 nucleotides were tested. Uncropped gel images are available in Supplementary Figure 1.