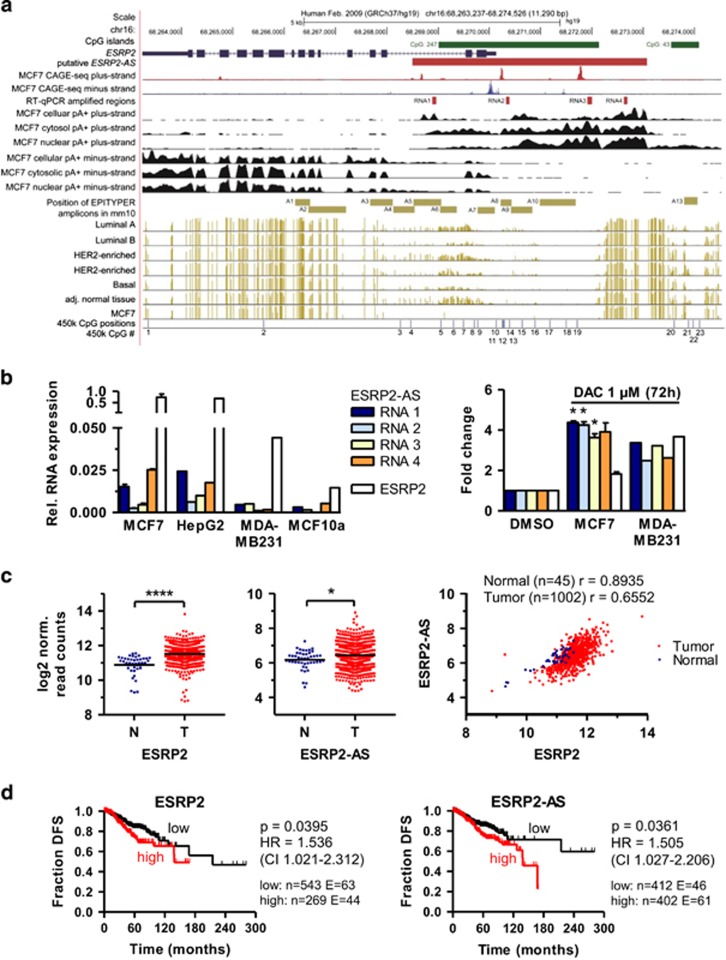
Figure 6.
A human homolog of Esrp2-as is overexpressed in human breast cancer and associated with poor prognosis. (a) UCSC Genome Browser40 scheme of the human ESPR2 locus on chr16 depicts the genomic location of the human ESRP2 gene (blue), CpG islands (green), genomic location of ESRP2-AS (red), MCF-7 CAGE-seq peaks for the positive (red) and negative strand (red), positions of RT–qPCR primers to quantify expression of ESRP2-AS (RNA1-4, red), strand-specific RNA-seq signals obtained with MCF7 whole cell lysates, cytosolic and nuclear fraction (black), location of mouse EPITYPER MassArray amplicons (using UCSC genome browser liftover tool, beige), DNA methylation levels for individual CpG sites (indicated by beige lines) assessed by WGBS (downloaded from TCGA), CpG sites covered on the Illumina 450 k methylation array (blue). (b) Left: Relative expression of ESRP2-AS (using four primer pairs designated as RNA1-4) and ESRP2 in MCF7 (n=2), HepG2 (n=1), MDA-MB231 (n=1) and MCF10a (n=1) human (cancer) cell lines, assessed by RT–qPCR. Right: Fold change of ESRP2 and ESRP2-AS levels in MCF7 (n=2) and MDA-MB231 cells (n=1) after DAC treatment, relative to DMSO solvent control set as 1. Statistical significance was assessed using the one-sample t-test with *P <0.05. (c) ESRP2 (left) and ESRP2-AS (middle) expression in human breast cancer samples from the TCGA BRCA breast invasive carcinoma dataset. RNA-seq data (log2 normalized read counts) for 1002 tumor samples were compared to 45 normal control samples by two-sided Student’s t-test with ****P <0.0001, *P<0.05. Right: Spearman correlation of ESPR2 and ESRP2-as levels (log2 norm. read counts) in normal breast tissue (blue) and tumor tissue (red) in the TCGA BRCA dataset. (d) Kaplan–Meier curves of disease-free survival are plotted for ESRP2 and ESRP2-AS expression. A total of 814 informative breast cancer cases from TCGA BRCA with an overall number of 107 events are separated by expression levels into a low (black) and high (red) expressing group. P-values for log-rank Mantel-Cox test; HR (hazard ratio) with 95% confidence interval indicated in parenthesis.