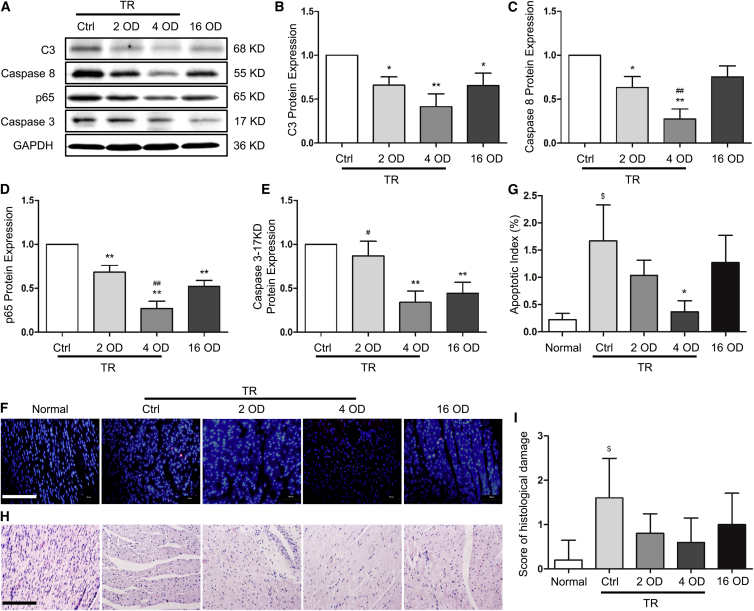
Figure 1.
siRNA Efficiency, Cardiac Tissue Survival, and Structure in Grafts Preserved with or without Transfection Reagent
Donor hearts were isolated, perfused and preserved (4°C) using transfection reagent (TR)-containing Celsior solution with 4 OD scramble siRNA (control group, Ctrl) or with 2 OD and 4 OD siRNA, or TR-free Celsior solution with 16 OD siRNA. 24 hr after preservation, tissue samples were collected. Fresh myocardium was used as a control (Normal). (A–E) siRNA efficacy was detected by western blot (A), and the expression of complement 3 (C3) (B), caspase-8 (C), p65 (D), and cleaved caspase-3 (17 KD) (E) was quantified by normalizing to GAPDH and relative to the Ctrl (set as 1). (F) Cardiac tissue survival was detected by TUNEL staining. (G) The apoptotic index was calculated by counting the apoptotic (red) and total cells in 10 fields at 400× magnification and is shown as the percentage of apoptotic cells (red) in total cells in each group. (H) Myocardial structural alteration was examined by H&E staining. (I) The semiquantitative score of histopathological damage was assessed in H&E sections in 5 fields at 400× magnification. 1 OD = 33 μg. Scale bars, 200 μm. Data shown are mean ± SEM. *p < 0.05, **p < 0.01 versus Ctrl; #p < 0.05, ##p < 0.01 versus the 16 OD group; $p < 0.05 versus Normal (n = 3).