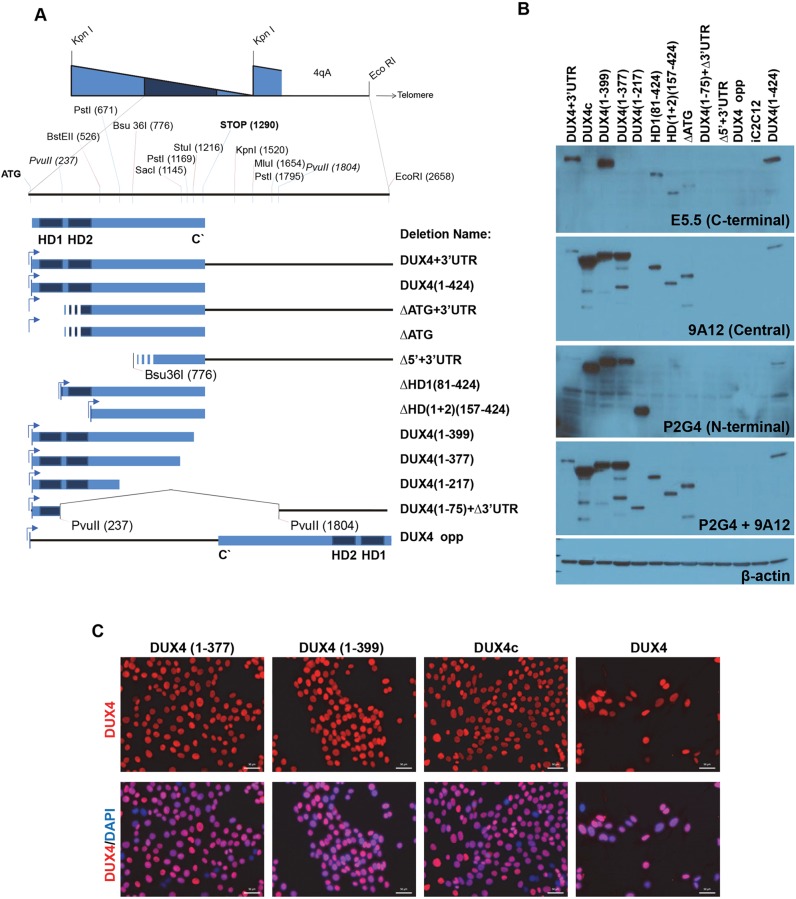
Fig. 1.
DUX4 deletion constructs and their inducibility. (A) Diagram of the DUX4 deletion constructs. 2.7 kb of DNA sequence containing the DUX4-ORF from the last full D4Z4 repeat, followed by its 3′UTR, was used as the template to generate deletions. Deletions were named with the amino acids of DUX4, listed as numbers in parentheses. HD denotes homeodomain. ‘DUX4’ used elsewhere in the manuscript may refer to either DUX4-ORF or DUX4+UTR. Stripes at the 5′ of some constructs indicate an unknown start signal as they lack the DUX4 ATG. (B) Western blotting demonstrates the expression and relative mass of each construct after 12 h of induction with 250 ng ml−1 of doxycycline. Protein expression was detected by DUX4-specific antibodies recognizing the N-terminus (P2G4), central (9A12) or C- terminus (E5-5) of DUX4; β-actin levels are also shown. (C) Immunostaining with DUX4 antibody (RD247c) and DAPI (nuclei, blue) of cells induced for 20 h with 250 ng ml−1 doxycycline in selected cell lines. Note that almost all of the nuclei are positively stained. Scale bars: 50 µm.