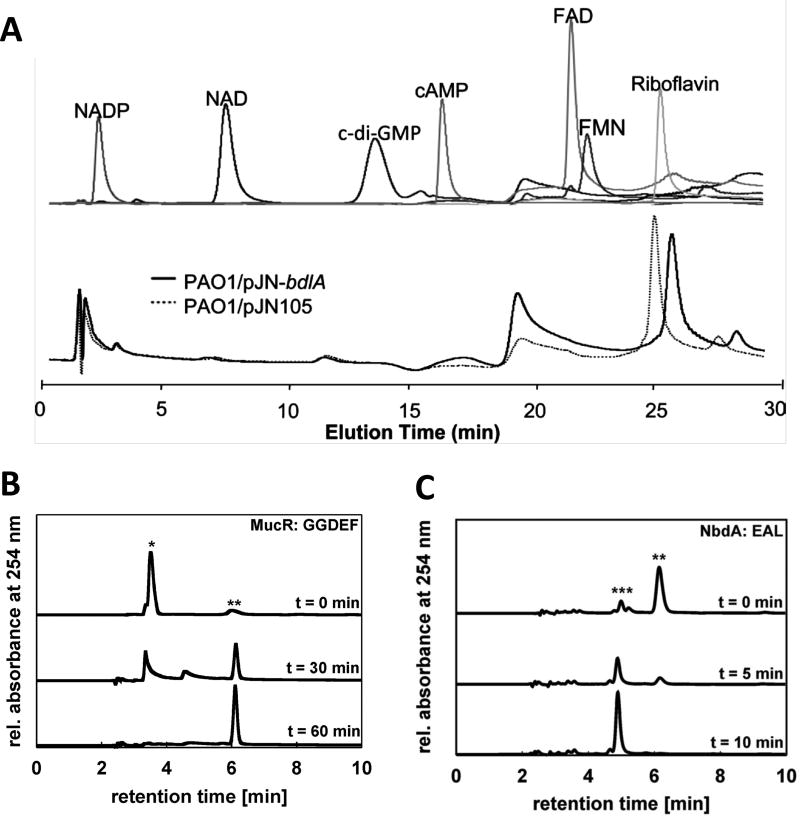
Figure 1. Separation of c-di-GMP and other small molecules by HPLC.
(A) Experiment demonstrating that the Pseudomonas aeruginosa chemotaxis protein BdlA does not bind FAD, FMN, NAD, NADP, cAMP, c-di-GMP or riboflavin. Top panel demonstrates the HPLC elution profiles of FAD, FMN, NAD, NADP, cAMP, c-di-GMP or riboflavin. Bottom panel demonstrates chromatograms of supernatants of purified BdlA (from P. aeruginosa PAO1/pJN-bdlA) or from vector control extracts (PAO1/pJN105). Reproduced from (22) with permission from American Society for Microbiology (ASM). (B) HPLC chromatograms demonstrating c-di-GMP synthesis by P. aeruginosa MucR. Peaks representing GTP (*) and c-di-GMP (**) are assigned. Reproduced from (17) with permission from ASM. (C) HPLC chromatograms showing c-di-GMP hydrolysis by the P. aeruginosa PDE NbdA. Peaks representing c-di-GMP (**) and pGpG (***) are assigned. Reproduced from (17) with permission from ASM. For all experiments, separation was performed on a C18 reversed-phase column, with analytes monitored at 254 nm.