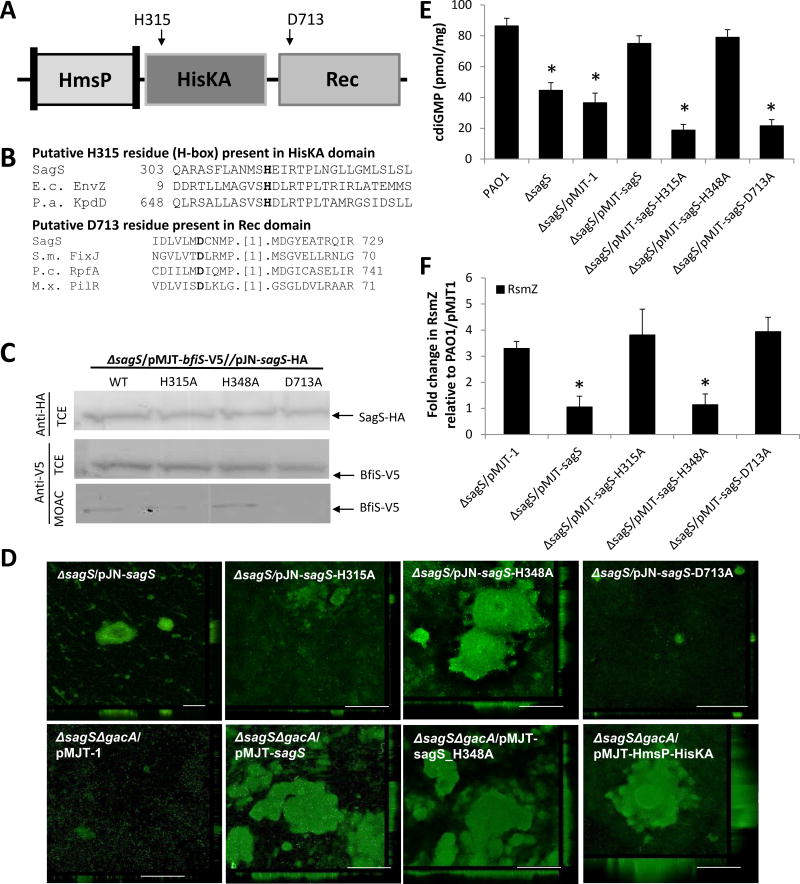
Figure 4. Conserved phosphotransfer residues of SagS contribute to biofilm development, but not antimicrobial tolerance.
(A) Overview of SagS conserved phosphorelay sites and corresponding site-directed mutagenesis targets. (B) Alignments showing the presence and location of invariant histidine and aspartate phosphorylation sites in the HisKA and Rec domains, respectively. Alignments were obtained via conserved domain BLAST; E.c. EnvZ, Escherichia coli EnvZ; P.a. KpdD, Pseudomonas aeruginosa two-component sensor KdpD; S.m. FixJ, Sinorhizobium meliloti transcriptional regulatory protein Fixj; P.c. RpfA, Pectobacterium carotovorum sensor/regulator protein RpfA; M.x. PilR, Myxococcus xanthus regulator protein PilR. (C) Detection of SagS and BfiS in total cell extracts (TCE) or MOAC-enriched phosphoproteomes (MOAC) of P. aeruginosa ΔsagS expressing wild-type sagS or sagS variants harboring alanine substitutions at amino acid residues H315, H348, and D713A by immunoblot analysis. Cell-free total cell extracts were obtained from P. aeruginosa grown biofilm cells. (D) Representative confocal images showing the architecture of biofilms formed by ΔsagS mutant strains expressing intact sagS or sagS variants harboring alanine substitutions at amino acid residues H315, H348, and D713A and the double mutant ΔsagSΔgacA harboring the empty plasmid pMJT-1 or pMJT containing sagS, sagS_H348A, or the SagS domain construct HmsP-HisKA. Biofilms were stained with the LIVE/DEAD BacLight viability stain. White bars = 100 µm. (E) c-di-GMP levels present in wild-type and ΔsagS mutant biofilm cells overexpressing sagS variants. The strains were grown in tube reactors under flowing conditions for 6 days prior to c-di-GMP extraction and quantitation by HPLC analysis. pmol/mg refers to c-di-GMP levels (pmol) per total cell protein (in mg). ΔsagS harboring the empty vector pMJT-1 was used as vector control. (F) RsmZ transcript levels as determined using qRT-PCR in cells grown as biofilms for 3 days. All experiments were carried out at least in triplicate. Error bars indicate standard deviation. *, significantly different from the values for P. aeruginosa PAO1 (p ≤ 0.01), as determined by ANOVA and SigmaStat.