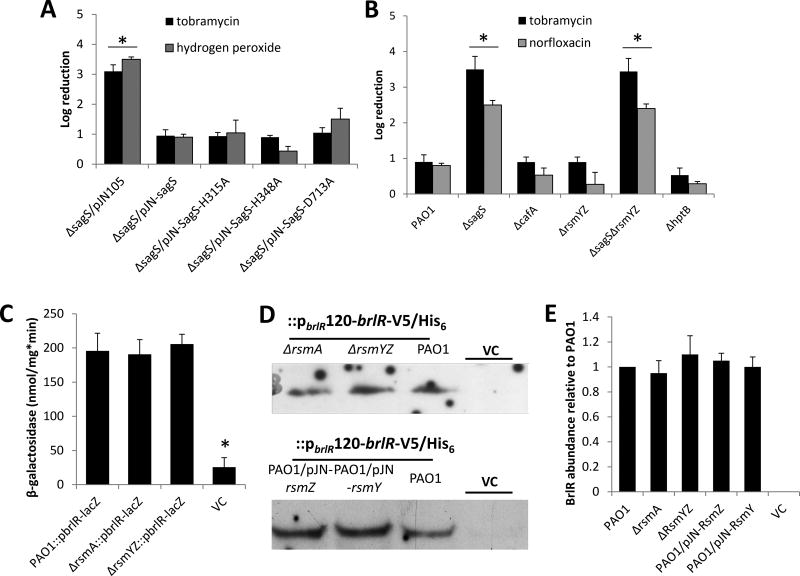
Figure 5. Susceptibility of biofilm cells and BrlR production is not linked to the phospho-transfer to BfiS, BfiS downstream targets or components previously linked to biofilm development.
(A–B) Susceptibility to tobramycin, hydrogen peroxide, or norfloxacin of biofilm cells of PAO1 and strains inactivated in cafA, rsmYZ, hptB, sagS and ΔsagS mutant strains expressing SagS variants. Biofilm susceptibility was determined as CFU log reduction. (C) Detection of brlR reporter activity in cells grown as biofilms for 3 days as determined using the chromosomal brlR-lacZ reporter construct and β-galactosidase activity. The promoter activity is reported as nmol ONPG cleaved per total cellular protein per minute. (D) Detection of BrlR in total cell extracts obtained from biofilm cells of P. aeruginosa PAO1 strains inactivated in RsmA or RsmYZ or overexpressing RsmY or RsmZ. All strains harbored a chromosomally located V5/His6-tagged BrlR under the control of its own promoter (PbrlR120-brlR-V5/His6), and respective protein extracts were probed for the presence of BrlR by immunoblot analysis using anti-V5 antibodies. A total of 15µg total cell extract was loaded. P. aeruginosa PAO1 harboring an empty CTX vector (VC) was used as negative control. Representative images are shown. (E) Relative BrlR abundance, based on relative intensity of protein bands detectable following probing for BrlR with anti-V5 antibodies and subsequent analysis using ImageJ (Rasband, 1997). Experiments were carried out in triplicate. Error bars denote standard deviation. *, significantly different from the values for P. aeruginosa PAO1 (p ≤ 0.01), as determined by ANOVA and SigmaStat.