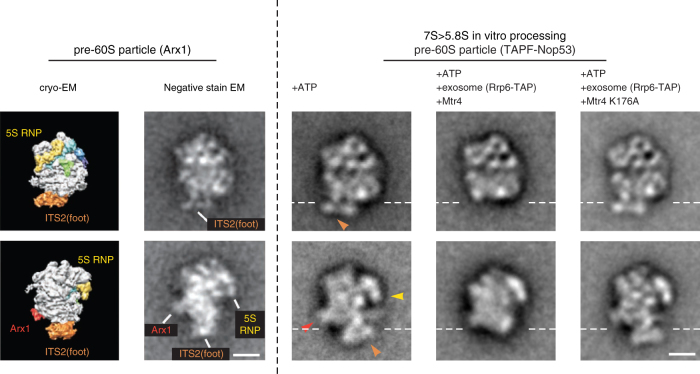
Fig. 4.
Detection of 'foot' structure removal from pre-60S particles. Negative-stain EM of the pre-60S particles (affinity-purified via TAPF-Nop53) after the in vitro processing reaction using endogenous yeast exosome (affinity-purified via Rrp6-FTpA) and recombinant wt Mtr4 or mutant Mtr4 (K176A), all in the presence of ATP. For control purposes, the pre-60S particle was also incubated with only ATP (mock). Shown to the left of the dashed line is the cryo-EM and negative-stain EM structure of the published Arx1-TAP particle (reproduced from ref. 25) in two orientations, rotated by 90° around the z-axis25, 26, which reveal structural hallmarks of the 7S pre-rRNA containing pre-60S particle: 'foot' (orange; ITS2 part of 7S pre-rRNA plus ITS2-associated factors), 5S RNP (yellow), Arx1 (red). Shown on the right are the two typical negative-stain EM classes of the pre-60S particles after the in vitro processing reaction, which correspond to the shown classes of the Arx1 particle. The pre-60S particles shown were derived from fraction 8 of the glycerol gradient (Fig. 3a). The orange, yellow and red arrowheads point to ITS2 (foot), 5S RNP and Arx1 structures, respectively. The white dashed horizontal lines mark where the 'foot' structure was removed from the core of the pre-60S particle upon in vitro 7S→5.8S pre-rRNA processing. The scale bars represent 10 nm. See Supplementary Fig. 4 for more class averages