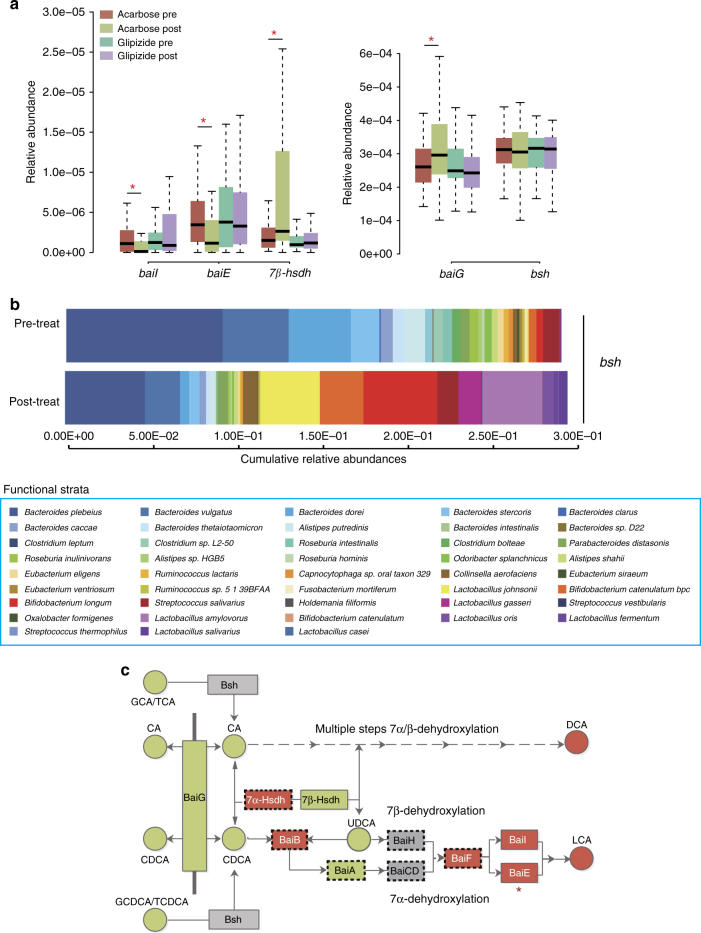
Fig. 4.
Acarbose treatment affects the potential for secondary bile acid metabolism. a Comparison of the relative abundances of genes encoding enzymes involved in SBA metabolism exhibiting significant alterations in abundance after Acarbose treatment, but not after Glipizide treatment; Plotted boxes are interquartile ranges. Dark lines in the boxes indicate medians, the lowest and highest values within 1.5 times IQR from the first and third quartiles, paired Wilcoxon rank-sum test, *p < 0.01, +p < 0.05. n = 51 in Acarbose pre and Acarbose post-treatment, n = 43 in Glipizide pre and Glipizide post-treatment. baiG, baiI and baiE, bile acid-inducible (bai) gene G, I and E; hsdh, gene encoding hydroxysteroid dehydrogenase, bsh, gene encoding bile salt hydrolase. b Cumulative relative abundances of genes encoding Bsh (EC.3.5.1.24) listed according to the contribution by annotated bacterial species pre-Acarbose and post-Acarbose treatment. The x-axis represents the cumulative relative abundances of genes encoding Bsh. Only species where the genes encoding Bsh constituted more than 0.5% of the total abundance of these genes, and where we observed a significant difference in relative abundances of these genes in response to Acarbose treatment, are included, q < 0.05, paired Wilcoxon rank-sum test. c Schematics of the bacterial BA biotransformation pathways, including deconjugation and multiple steps of 7α/β-dehydroxylation. Faecal BAs and BA-metabolising enzymes encoded by genes enriched after Acarbose treatment are marked in green, those depleted after Acarbose treatment are marked in red; those that did not change in abundance by treatment are marked in grey. *indicates the rate-limiting enzyme of 7α-dehydroxylation. Bsh, bile acid hydrolase; Bai G/B/A/I/H/CD/E/F, enzymes encoded by the bai operon. GCA glycocholic acid, TCA taurocholic acid, GCDCA glycochenodeoxycholic acid, TCDCA taurochenodeoxycholic acid, CA cholic acid, CDCA chenodeoxycholic acid, UDCA ursodeoxycholic acid, DCA deoxycholic acid, LCA lithocholic acid. Boxes framed by dashed lines represent enzymes where the annotation is ambiguous because of inconsistent functional annotation using BlastP based on the Uniprot database vs. BlastKOALA based on the KEGG database. (See Supplementary Methods, Functional annotation of genes involved in BA synthesis based on BLASTP and BlastKOALA for further details)