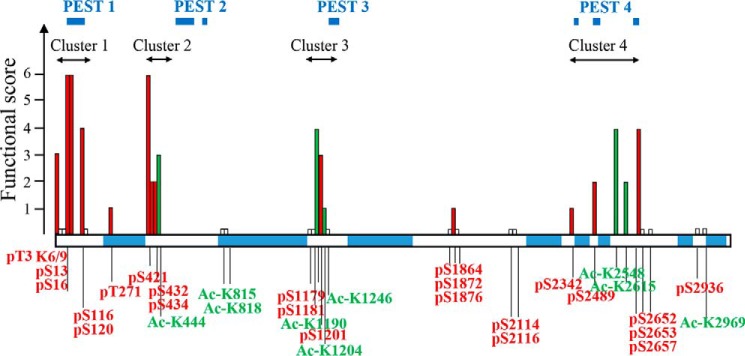
Figure 5.
Functional assessment of PTMs on Htt. PTMs clustering within proteolytic domains modulate mutant Htt toxicity. The functional score has been calculated for each PTM, based on the outcome of all the assays (toxicity in cortex, toxicity in striatum, mitochondrial potential, and mitochondrial size). Each PTM was given a score of 1 per assay if significant changes were observed and 0 if there was no difference. For multiple alterations of the same amino acid, a combined score was recorded. Ser13 and Ser16 are scored based on the single and combined alterations results. Based on this scoring, we found four clusters of PTMs that largely modulate expanded Htt functional or toxic properties. These clusters are located within protease-sensitive (PEST) regions (predicted by epestfind-EMBOSS Explorer) as indicated. Phosphorylation sites are shown in red, acetylation sites are in green, and sites that did not score in any of the assays are indicated by short white bars.