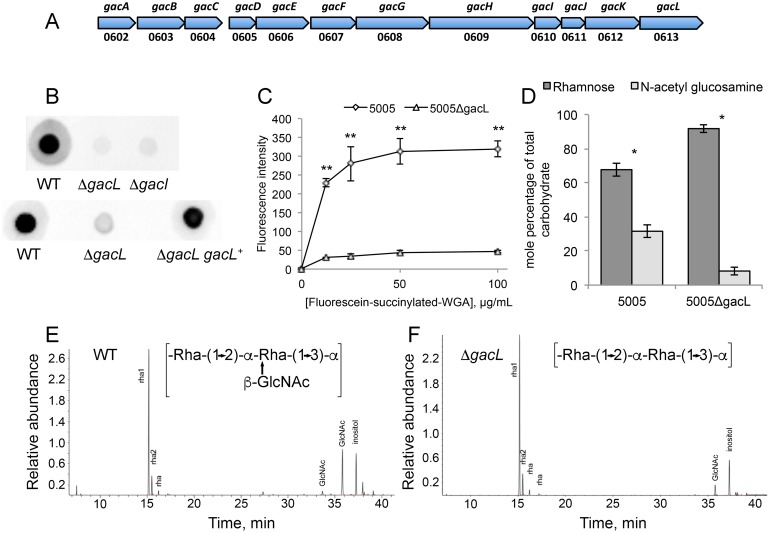
Figure 1.
Map of S. pyogenes genes involved in GAC biosynthesis and analysis of 5005ΔgacI and 5005ΔgacL deletion mutants. A, GAC biosynthesis gene cluster. Numbers below represent MGAS5005 gene designations. B, representative immunoblot analysis of cell-wall fractions isolated from MGAS5005, 5005ΔgacI, 5005ΔgacL, and 5005ΔgacL gacL+. Data are representative of biological triplicates. C, binding of N-acetylglucosamine-specific fluorescein-succinylated WGA to whole MGAS5005 and 5005ΔgacL was measured. Data are the average of three replicates ± S.D. D, rhamnose and GlcNAc mole percentage of total carbohydrate was determined by GC-MS for cell wall material isolated from MGAS5005 and 5005ΔgacL following methanolysis as described under “Experimental procedures.” Data are the average of four replicates ± S.D. The asterisks indicate statistically different values (*, p < 0.05; **, p < 0.01) as determined by the Student's t test. E and F, GC-MS chromatograms for glycosyl composition analysis of cell wall isolated MGAS5005 and 5005ΔgacL. The deduced schematic structure of the repeating unit of GAC is shown for each strain. The chromatograms are representative of four separate analyses performed on two different cell wall preparations.