Figure 4.
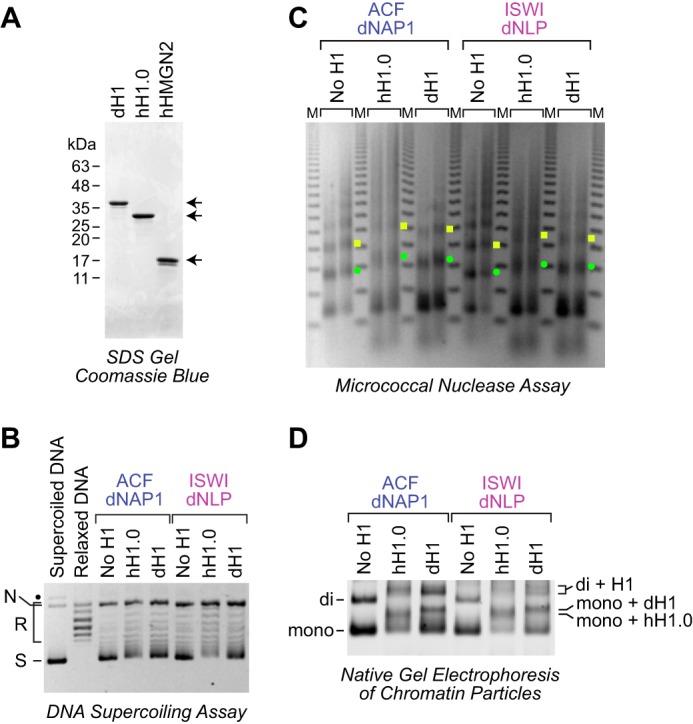
The dNLP-ISWI system can be used to assemble histone H1–containing chromatin. A, purification of native Drosophila histone H1, recombinant human H1.0, and recombinant human HMGN2. The proteins were subjected to 15% polyacrylamide-SDS gel electrophoresis and staining with Coomassie Brilliant Blue R-250. B, DNA supercoiling analysis of chromatin assembled with histone H1. Reactions were performed either with the dNAP1-ACF system (4) or with the dNLP-ISWI system, as described in the legend for Fig. 3. Where indicated, Drosophila histone H1 and human H1.0 were included at a ratio of 0.75 molecules of H1/nucleosome. The positions of nicked DNA (N), relaxed DNA (R), and supercoiled DNA (S) are shown. The black dot corresponds to a minor unknown contaminant. C, partial MNase analysis shows an increase in the nucleosome repeat length upon incorporation of histone H1 into chromatin. Reactions were performed as described in B, and the resulting samples were subjected to partial MNase digestion analysis. The green dots correspond to the DNA bands derived from dinucleosomes, and the yellow squares designate DNA fragments from trinucleosomes. D, native gel electrophoresis of mono- and dinucleosomes obtained by extensive MNase digestion of chromatin either lacking or containing histone H1. The chromatin particles were detected by staining of the DNA with SYBR Gold. The positions of mononucleosomes (mono), dinucleosomes (di), H1-containing mononucleosomes (chromatosomes; mono + H1), and H1-containing dinucleosomes (di + H1) are indicated.
