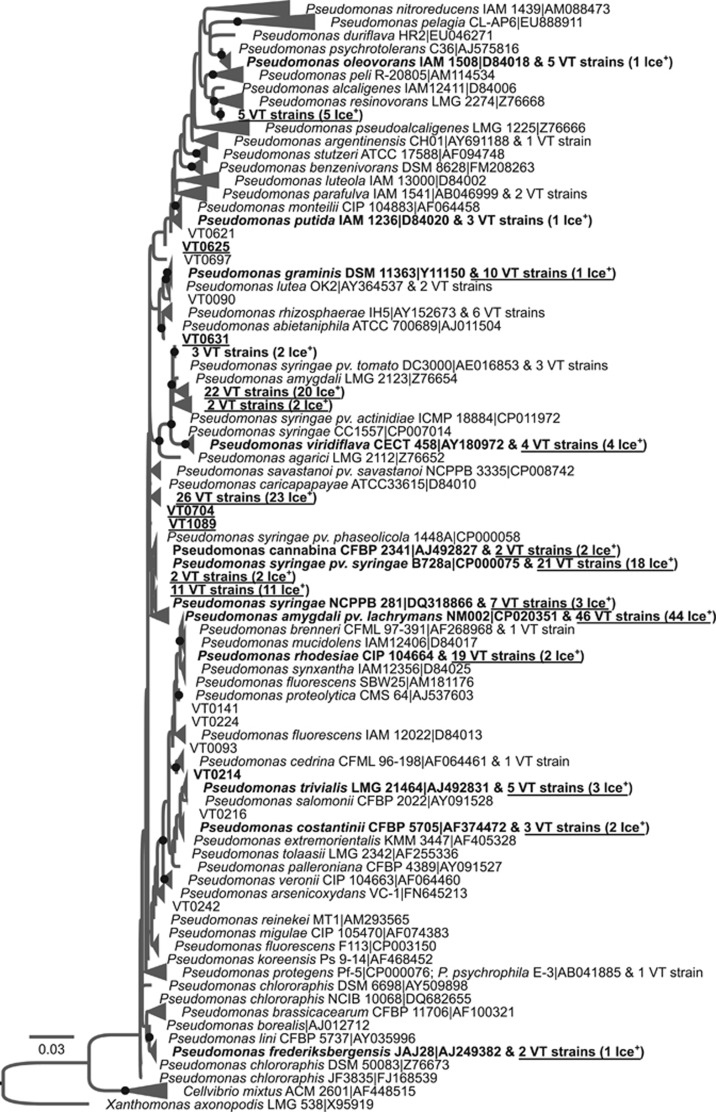
Figure 2.
Evolutionary relationships between 229 Ice+ strains isolated from precipitation in Virginia, USA (labeled 'VT#') and 151 type strains of the Pseudomonadaceae family based on the alignment of partial 930bp-long 16S rDNA sequences. A maximum likelihood tree was built using the GTR model and 100 bootstrap replicates. Gaps were considered as missing data and were partially deleted with a site coverage cut-off of 95%. The strain Xanthomonas axonopodis LMG 538-T was used as a root. Only bootstrap values over 50% were included and are symbolized by black dots. Strains that were Ice+ in standardized re-tests at −8 °C are labelled in bold and are underlined, those active at −10 °C or −12 °C in standardized re-tests are labelled in bold only, and those that were only active in the initial test but not in the re-tests are neither underlined nor in bold. When more than one VT# strain had the same 16S rDNA sequence, only the number of strains is indicated and Ice+ strains that were confirmed in repeated tests are labelled ‘Ice+’. RDP names are composed of the species name, the strain code and the accession number in the public database NCBI. The name of the type strain closest to Ice+ strains was chosen to represent collapsed branches. A version of the tree in which all VT strains are listed is given in Supplementary Figure S3.