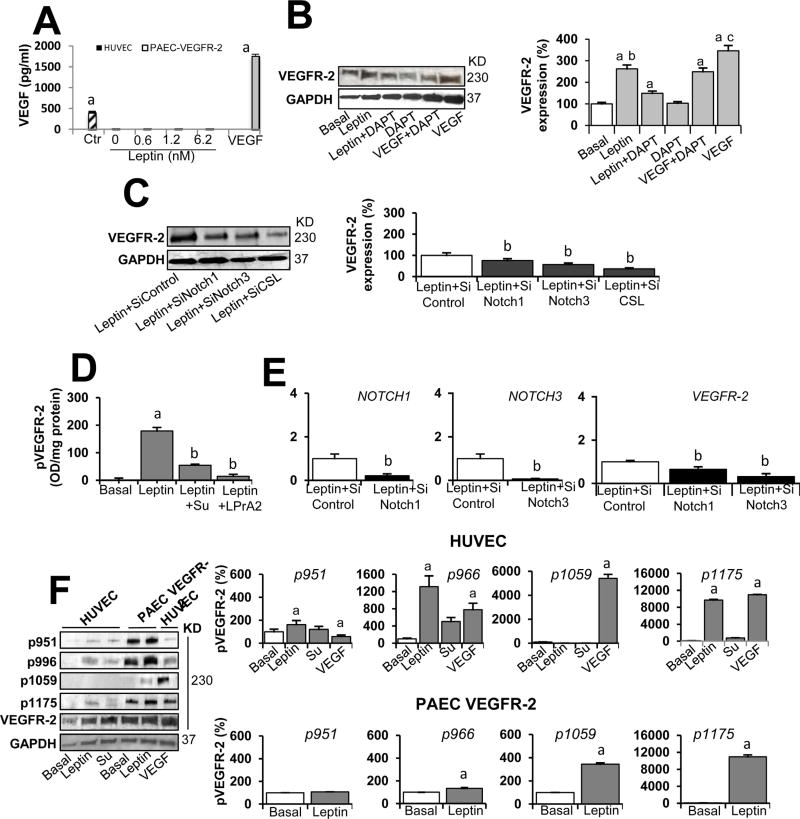
Fig. 7. Leptin-induced expression and phosphorylation of VEGFR-2 in endothelial cells is independent of VEGF.
A. VEGF levels in culture supernatant from endothelial cells treated with leptin. HUVEC and PAEC VEGFR-2 cells were cultured for 24 h with leptin (0, 0.6, 1.2, and 6.2 nM). VEGF165 levels (µg/ml) in cell culture supernatants were determined via ELISA (R&D System). ELISA kit control (Ctr) and VEGF (positive control) were also tested. Assay dynamic range and sensitivity were 15.6-1000 pg/ml and 5pg/ml, respectively. B. Leptin induction of VEGFR-2 protein in HUVEC is reduced by Notch inhibition. Represented here are western blot results from the effects of Notch inhibition on leptin induction of VEGFR-2. HUVEC were treated for 24 h with leptin [0 (basal) and 1.2 nM], Notch inhibitor (DAPT, gamma-secretase inhibitor; 5µmol/l). VEGF (25 ng/ml) and VEGF + DAPT. GAPDH was used as a loading control. C. Leptin induction of VEGFR-2 protein in HUVEC is reduced by Notch knockdown. WB representative results of VEGFR-2 from the effects of mRNA silencing of Notch and CSL. HUVEC wild type and transfected with Notch1, Notch3, and CSLSiRNA were treated for 24 h with leptin [0 (basal) and 1.2 nM]. Relative protein expression was calculated as percentage to basal. Histograms show densitometric analysis of Notch protein expression using NIH image J software. D. Leptin-induced phosphorylated VEGFR-2 (pVEGFR-2) levels in HUVEC is reduced by VEGFR-2 kinase inhibition. HUVEC were treated for 24 h with leptin [0 (basal) and 1.2 nM] and inhibitors of VEGFR-2 kinase (SU5416; 5 µmol/l) and leptin receptor (LPrA2; 1.2 nM). Levels of VEGFR-2 in cell lysates were determined by ELISA (R&D Systems). Assay dynamic range and sensitivity were 78.1-5,000 pg/ml and 11.4 pg/ml, respectively. E. Leptin induction of VEGFR-2 mRNA in HUVEC is reduced by Notch knockdown. Quantitative results from real-time PCR of VEGFR-2 and Notch1 and Notch3 mRNA expression in HUVEC treated with leptin and Notch1 and Notch3 SiRNA. RNA expression was calculated by normalizing values to GAPDH mRNA. Relative mRNA expression was calculated to basal. F. Leptin induction ofVEGFR-2 phosphorylation in endothelial cells. WB representative results from leptin-induced phosphorylation of VEGFR-2 in HUVEC and PAEC VEGFR-2 in absence of VEGF. Cells were incubated with leptin (0 and 1.2 nM) and VEGFR-2 kinase inhibitor (SU5416; 5 µmol/l) for 24 h. VEGF (25 ng/ml) was used as a positive control. Levels of phosphorylated VEGFR-2 (pVEGFR-2 at Y951, Y966, Y1059 and Y1175) were determined by WB using specific antibodies. Histograms show densitometric analysis of pVEGFR-2 proteins using NIH image J software. GAPDH was used as a loading control. Relative protein expression was calculated as percentage to basal. Data is presented as an average ± s.d. from three independent experiments. Su: SU5416; SiControl: siRNA control. a: p<0.05 when compared to basal. b: p< 0.05 when compared to endothelial cells treated with leptin.