Figure 1.
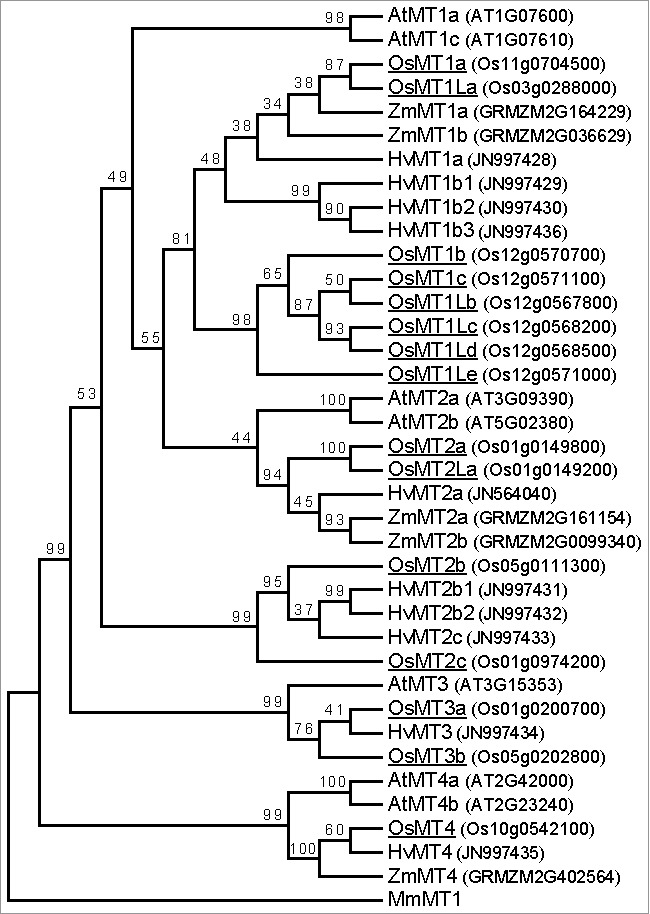
Phylogenetic analysis of MT proteins in plants. The amino acid sequences of the MT proteins were aligned by the ClustalW (MEGA6 package). MEGA6 (http://www.megasoftware.net) was used for construction of the neighbor-joining phylogenetic tree with bootstrap values calculated based on 1000 replicates. MT proteins from monocotyledonous plants [rice (Oryza sativa; Os), maize (Zea mays ssp. mays; Zm), and barley (Hordeum vulgare; Hv)], and dicotyledonous plants [Arabidopsis (Arabidopsis thaliana; At)] were used for the analysis. The rice OsMT proteins were indicated by underlines. MmMT1 from mouse (Mus musculus) was used as the out-group. Accession numbers or IDs of each protein were denoted in the parentheses. The amino acids sequences were obtained from each database as described by Yamauchi and colleagues.10
