Fig. 1.
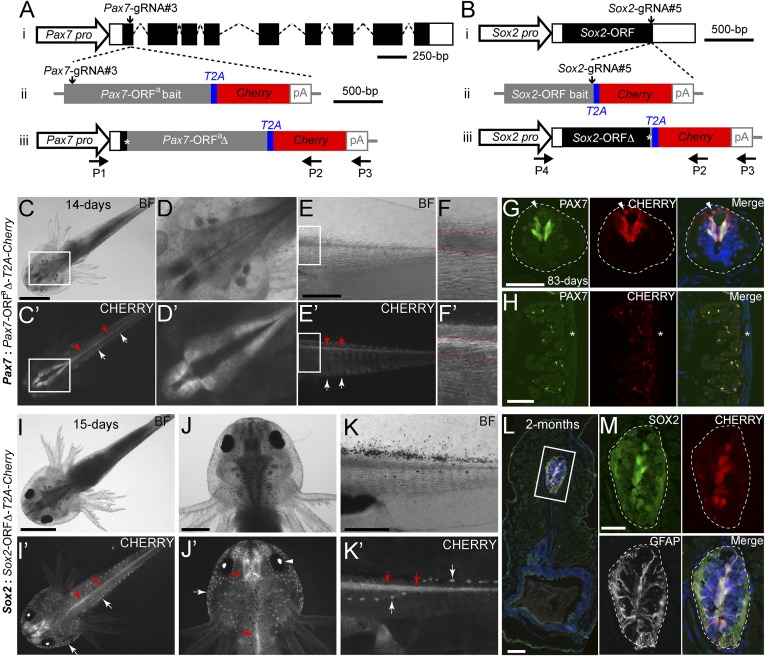
Knockin of a Cherry reporter gene into two axolotl genomic loci through CRISPR/Cas9- mediated homologous-independent integration. (A and B) The knockin strategies for the generation of the Pax7: Pax7-ORFa∆-T2A-Cherry (A) and Sox2: Sox2-ORF∆-T2A-Cherry (B) axolotl reporter lines. (i) The wild-type axolotl Pax7 (A, Dataset S1) and Sox2 (B) gene structures. Solid rectangles represent coding sequences of the exons; empty rectangles, untranslated regions; dashed lines: introns. The gRNA targeting sites are indicated by vertical arrows. (ii) The targeting constructs, pGEMT-Pax7-ORFa-T2A-Cherry-PA (A) and pGEMT-Sox2-ORF-T2A-Cherry-PA (B), contain the entire axolotl Pax7 (A; Pax7-ORFa bait) or Sox2 (B; Sox2-ORF bait) ORF of the cDNA (with the start codon, but without the stop codon) as the bait sequence, followed by T2A, the Cherry coding sequence, and the polyadenylation signal (pA). Vertical arrows indicate the gRNA targeting sites. (iii) The Pax7 (A) and Sox2 (B) alleles after knockin of the Cherry reporter gene. Asterisks indicate the junctions after the integration of the targeting constructs. The newly formed mosaic Pax7 (A) and Sox2 (B) coding sequences are designated Pax7-ORFaΔ and Sox2-ORFΔ, respectively. The horizontal arrows indicate the binding positions of the genotyping primers (P1–P4). (C–F′) Bright-field (BF; Upper) and CHERRY fluorescence (Lower) images of 14-d-old Pax7: Pax7-ORFaΔ-T2A-Cherry knockin F0 axolotls. The dorsal (C–D′) and lateral (E–F′) view images highlight the CHERRY expression in the brain (C–D′), spinal cord (red arrows), and trunk muscle (white arrows) compartments. The areas depicted by rectangles in C and C′ and E and E′ are shown at higher magnification in D and D′ and F and F′, respectively. The red dashed lines (F and F′) define the spinal cord region showing that the CHERRY expression is restricted in the dorsal domain of the spinal cord. (Scale bars: 2 mm in C, 1 mm in E.) (G and H) Immunofluorescence for PAX7 (green), CHERRY fluorescence (red), and DAPI (blue) on 10-μm tail cross-cryosections of 83-d-old Pax7: Pax7-ORFaΔ-T2A-Cherry knockin F0 axolotls shows that CHERRY expression is restricted to the PAX7-expressing domain in dorsal spinal cord (G, dashed line circles) and satellite cells (H). The arrowhead indicates inherited CHERRY in a PAX7-negative newborn neuron. The asterisk indicates tail skin. (Scale bars: 100 μm in G and H). (I–K′) Bright-field (BF, Upper) and CHERRY fluorescence (Lower) images of 15-d-old Sox2: Sox2-ORFΔ-T2A-Cherry knockin F0 axolotls. The dorsal view (I–J′) and lateral view (K and K′) images highlight the CHERRY expression in the central nervous system (red arrows), the lens (arrowhead), and the lateral line neuromasts (white arrows). (Scale bars: 2 mm in I, 1 mm in J and K.) (L and M) Immunofluorescence for SOX2 (green), GFAP (white), and CHERRY fluorescence (red) combined with DAPI (blue) on 10-μm cross-cryosections of 2-mo-old Sox2: Sox2-ORFΔ-T2A-Cherry knockin F0 axolotls shows that CHERRY expression is restricted to SOX2 positive cells in the spinal cord (dashed circles) (M). GFAP is shown to highlight the morphology of the luminal SOX2-positive radial glial cells expressing GFAP. The rectangle depicted in L is shown as separated or merged images at higher magnification in M. (Scale bars: 100 μm in L, 50 μm in M.)