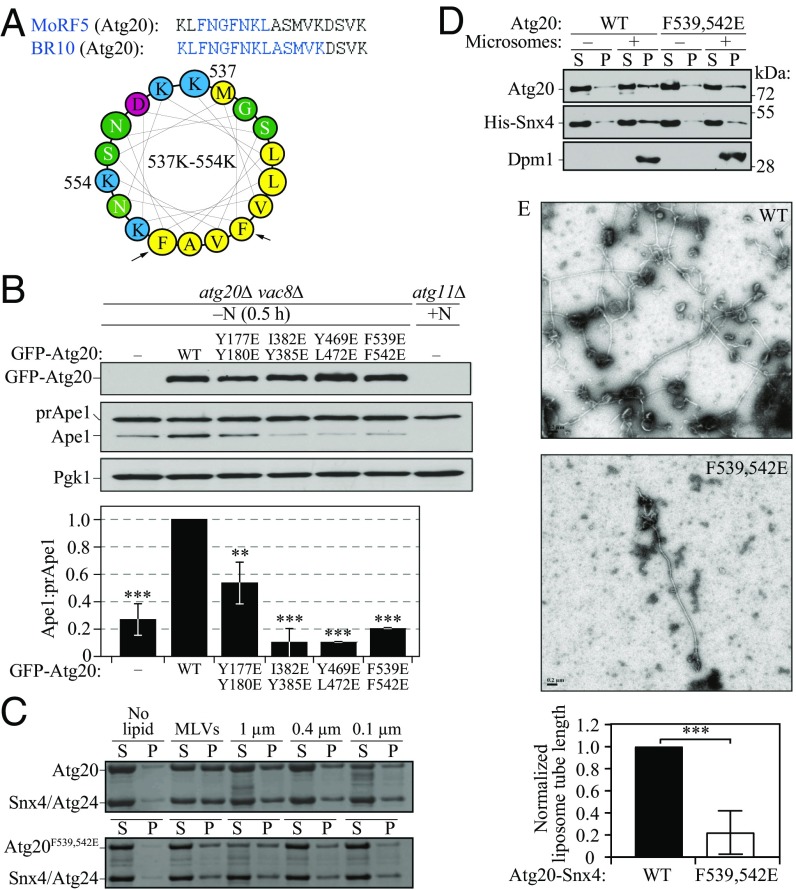
Fig. 6.
The membrane-induced AH in Atg20. (A) Helical wheel representation of the amino acid sequence in Atg20 that corresponds to MoRF5/BR10. Black arrows indicate the double mutation F539E, F542E. Yellow indicates hydrophobic amino acid residues; green, polar; red, negatively charged; blue, positively charged. (B) The vac8∆ atg20∆ (HPY079) cells transformed with the plasmids {pCuGFP(426), pCuGFP-Atg20(426), pCuGFP-Atg20[Y177E Y180/E](426), pCuGFP-Atg20[I382E Y385E](426), pCuGFP-Atg20[Y469E L472E](426), and pCuGFP-Atg20[F539E F542E](426)} were cultured in rich selective medium and then nitrogen-starved for 0.5 h. Error bars represent SD from three independent experiments. (C) Liposome sedimentation assay for the recombinant Atg20-Snx4 heterodimer, in which Atg20 was either the WT or F539E F542E mutant, with Folch liposomes of varying diameter. (D) In vitro reconstitution of yeast microsomes, isolated from SEY6210 atg20Δ snx4Δ cells, with the recombinant purified heterodimer Atg20-Snx4, in which Atg20 was either WT or mutant including the F539E F542E mutation. Supernatant (S) and pellet (P) were obtained by ultracentrifugation and analyzed by western blot analysis. (E, Upper) Representative negative stain EM images of 1.0-μm vesicles incubated with WT Atg20-Snx4 or mutant Atg20-Snx4 [F539E F542E] heterodimer. (Scale bars: 0.2 µm.) (E, Lower) Quantification of lipid tube length from grid squares, which are 484 µm2. Data from each trial were normalized to the tube length from WT. Error bars represent the SD from three independent experiments. Statistical significance was tested using the unpaired two-tailed Student’s t test: **P < 0.01; ***P < 0.005.