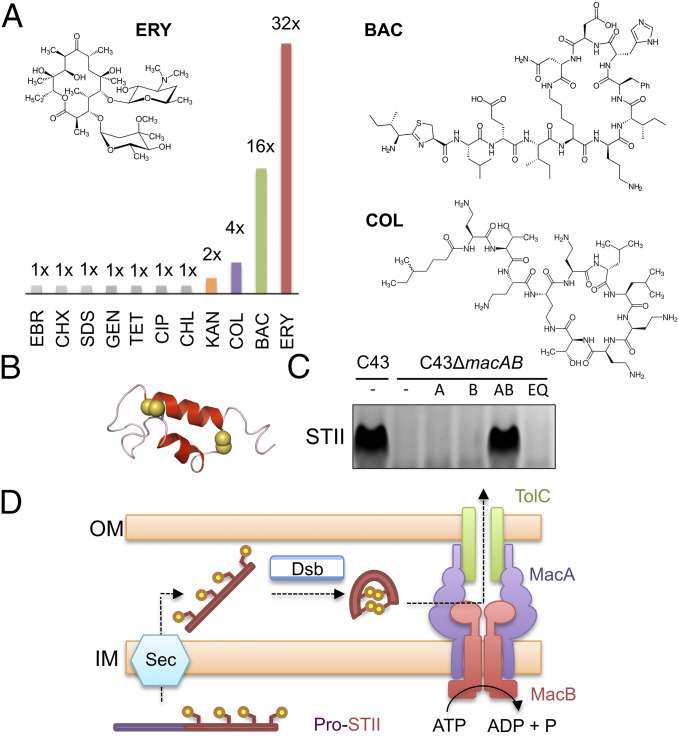
Fig. 2.
Functional analysis of E. coli MacAB-TolC in vivo. (A) Antibiotic susceptibility comparison between E. coli C43 (DE3) ΔacrABΔmacAB cells expressing E. coli MacA and either wild-type or Glu170Gln E. coli MacB variant. The ratio of the MIC for the wild-type MacB to the Glu170Gln variant is shown graphically for ethidium bromide (EBR), chlorhexidine (CHX), SDS, gentamycin (GEN), tetracycline (TET), ciprofloxacin (CIP), chloramphenicol (CHL), kanamycin (KAN), colistin (COL), bacitracin (BAC), and erythromycin (ERY). Structures of erythromycin, bacitracin, and colistin are shown (Inset). (B) Structure of enterotoxin STII with disulfide bond-forming sulfur atoms (yellow spheres; Protein Data Bank: 1EHS) (32). (C) Detection of secreted enterotoxin in E. coli culture supernatants by SDS/PAGE. Lanes from left to right: C43 wild-type, C43ΔmacAB, C43ΔmacAB expressing plasmid-borne macA (A), macB (B), macAB (AB), or macAB with an ATPase inactivating Glu170Gln substitution (EQ). All strains contain a plasmid-expressing enterotoxin STII. (D) Model for two-step secretion of enterotoxin STII across the E. coli cell envelope. Sec and Dsb represent the general protein secretion machinery and disulfide bond incorporation systems, respectively.