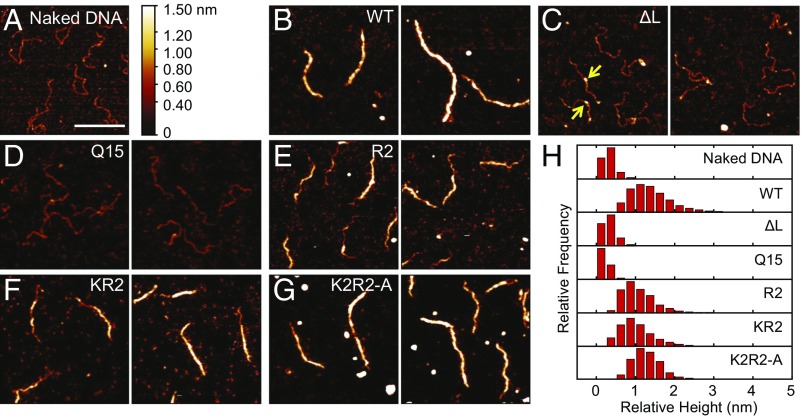
Fig. 2.
DNA binding is impaired in H-NS linker mutants. (A) The csgD promoter in the absence of H-NS protein. (B) Wild-type H-NS binds and polymerizes along the DNA, forming a stiffened filament. Linker mutants lacking the linker (C) or with the polyQ linker (D) form small foci (yellow arrows) due to reduced binding affinity (SI Appendix, Fig. S3) and fail to polymerize. Addition of positive charges improves binding, leading to DNA polymerization in E and F. The K2R2 mutant (G) binds similarly to the wild-type protein. Two panels show different representative images. All proteins were present at 600 nM, and binding was performed in 50 mM KCl, 2 mM MgCl2, 10 mM Tris⋅HCl (pH 7.4) buffer. (H) Relative height distribution histograms were obtained from DNA contours of each experiment. (Scale bar: 200 nm.)